CRISPR Plasmids: Prime Edit
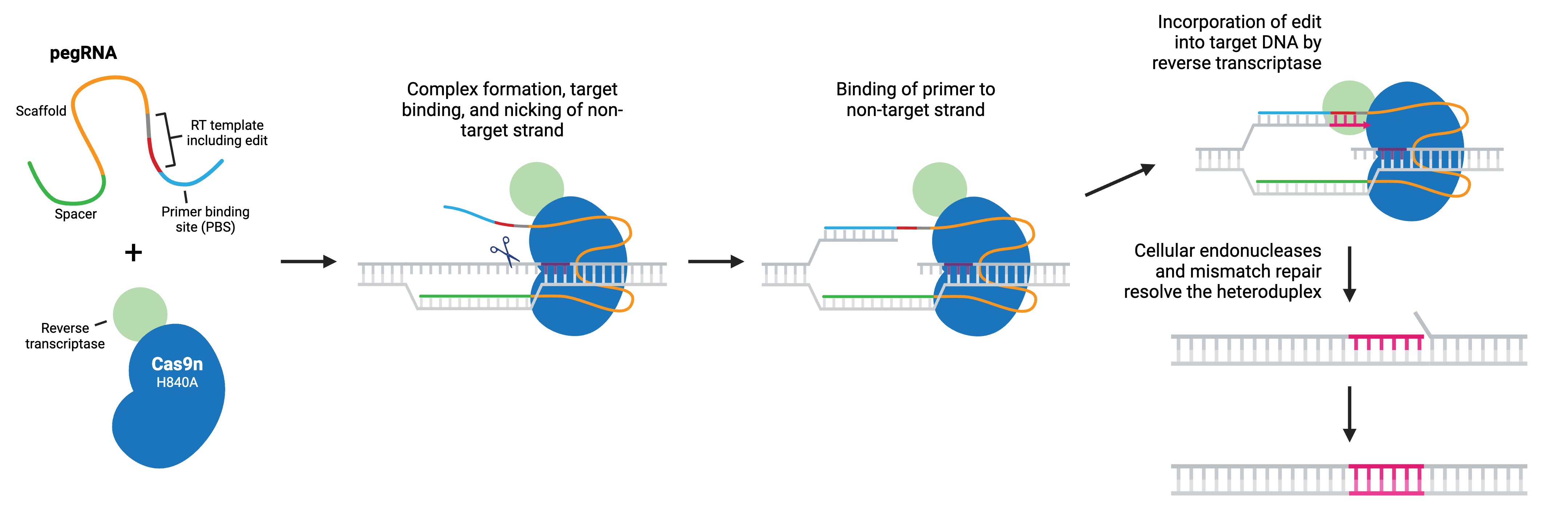
Prime editing is a “search and replace” gene editing method in which a reverse transcriptase (RT) is fused to the C terminus of Cas9 H840A nickase. The fusion enzyme is capable of installing targeted insertions, deletions, and point mutations using a prime editing guide RNA (pegRNA). As with a typical gRNA, the pegRNA is designed with a spacer that binds to a specific genomic DNA locus and directs the nickase to the target site. The longer pegRNA also encodes a primer binding site (PBS) and the desired edits on an RT template.
During prime editing, the pegRNA directs the Cas9 nickase to the target sequence where it nicks the non-target strand and generates a 3’ flap. The 3’ flap binds to the PBS of the pegRNA and the desired edit is incorporated into the DNA by reverse transcription. The edited DNA strand displaces the unedited 5’ flap and the resulting heteroduplex is resolved by the cell’s mismatch repair (MMR) system. Alternatively, if the edited 3’ flap is excised, the target sequence remains unchanged and available as a substrate for another round.
The original prime editing enzyme is named PE1. PE1 and most other prime editors use the Moloney murine leukemia virus (M-MLV) reverse transcriptase. Engineering of the PE enzyme and related components have helped to improve the efficiency of editing, including exploring new sources for the reverse transcriptase. Many PE enzymes and related tools are available:
- PE2 — introduction of five mutations in RT enzyme
- PE3 — PE2 plus additional sgRNA
- PE4 — PE2 plus MLH1 (a protein component of MMR) mutant to inhibit MMR
- PE5 — PE3 plus MLH1 mutant to inhibit MMR
- PEmax — optimized PE2 enzyme containing RT optimized for human codons, additional nuclear localization signals, and two mutations in Cas9 to enhance nuclease activity; can be used in PE2–PE5 systems
- PE6 — small prime editors with optimized RT and/or Cas9 domains
- PE7 — addition of RNA-binding exonuclease protection factor La to PEmax to enhance pegRNA stability
- epegRNA — additional protection added to 3’ tail of pegRNA to prevent RNA degradation
For more in-depth information on these PE tools, check out our prime editing blog post. Need help designing a pegRNA? Try the DeepPrime (Link opens in a new window) design tool from Hyongbum Kim's lab.
Browse, sort, or search the tables below for CRISPR prime editing plamisds. To learn more about prime editing and other CRISPR topics, read our CRISPR Guide.
Mammalian
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Bacteria
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Drosophila
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Plant
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Empty Prime Editing gRNA Vectors
A selection of empty gRNA vectors suitable for prime editing are highlighted in the table below. Use the search bar to find a gRNA vector based on expression system, promoter, the type of gRNA (e.g., pegRNA, epegRNA, nicking sgRNA), cloning enzyme, selectable marker, and whether the plasmid contains Cas9.
| ID | Plasmid | Expression System | Promoter | Guide RNA Type | Cloning Enzyme | Co-expressed Cas9 | Selection | PI | |
|---|---|---|---|---|---|---|---|---|---|
| 132777 | pU6-pegRNA-GG-acceptor | Mammalian | hU6 | pegRNA | BsaI | No | mRFP1 | David Liu | |
| 140448 | QPM-sgR (pTaU3) | Plant | TaU3 | nicking sgRNA | Eps3I + NcoI | No | Caixia Gao | ||
| 141081 | pYPQ141D-peg | Plant | OsU3 | pegRNA | No | Yiping Qi | |||
| 149545 | pCFD3-NS | Drosophila | Drosophila U6:3 | pegRNA | BbsI | No | Vermilion | Norbert Perrimon | |
| 149546 | pCFD5-NS | Drosophila | Drosophila U6:3 | pegRNA + nicking sgRNA | BbsI | No | Vermilion | Norbert Perrimon | |
| 164423 | pHSG1C3 | Mammalian | U6 | pegRNA + nicking sgRNA | BbsI for sgRNAs; BbsI + PstI for pegRNAs | No | Xiao Wang | ||
| 170132 | pOsU3 | Plant | OsU3 | pegRNA | BsaI + HindIII | No | Caixia Gao | ||
| 172716 | pPEgRNA | Bacteria | J23119 (BBa_J23119) | pegRNA | SpeI + HindIII | No | Tilmann Weber | ||
| 172717 | pnsgRNA | Bacteria | J23119 (BBa_J23119) | nicking sgRNA | No | Tilmann Weber | |||
| 173220 | pPBT-peRNA_GG-Puro | Mammalian, piggyBac | hU6 | pegRNA | BsmBI | No | Puromycin | Jacob Giehm Mikkelsen | |
| 173222 | pPBT-PE2-PuroTK-pegRNA_GG | Mammalian, piggyBac | hU6 | pegRNA | BsmBI | Yes (Cas9 H840A + MMLV RT) | PuroTK | Jacob Giehm Mikkelsen | |
| 174038 | pU6-tevopreq1-GG-acceptor | Mammalian | hU6 | epegRNA | BsaI | No | mRFP1 | David Liu | |
| 174039 | pU6-tmpknot-GG-acceptor | Mammalian | hU6 | epegRNA | BsaI | No | mRFP1 | David Liu | |
| 176901 | U6-pe RNA-H1-nick sgRNA-mCherry | Mammalian, AAV | hU6 + H1 | pegRNA + nicking sgRNA | No | mCherry | Hyongbum Kim | ||
| 177180 | pDAS12069_U6-pegRNA-mCherry | Mammalian | hU6 | pegRNA | BpiI for pegRNA, Esp31 for PBS-RT | No | mCherry | Ervin Welker | |
| 177181 | pDAS12222_U6-pegRNA-BFP | Mammalian | hU6 | pegRNA | BpiI for pegRNA, Esp31 for PBS-RT | No | BFP | Ervin Welker |
CRISPR Resources
Addgene has a large selection of CRISPR plasmids and resources. Find more CRISPR functions along with plasmids categorized by organism by visiting our CRISPR plasmids page. Find a comprehensive list of CRISPR resources by visiting our CRISPR reference page.
Content last reviewed: 17 October 2025
Do you have suggestions for other plasmids that should be added to this list?
Fill out our Suggest a Plasmid form or e-mail [email protected] to help us improve this resource!



