CRISPR Plasmids: Base Edit
Two classes of base editors — cytosine base editors (CBEs) and adenine base editors (ABEs) — can be used to generate single base pair edits without double stranded breaks.
Cytosine base editors are created by fusing Cas9 nickase or catalytically inactive “dead” Cas9 (dCas9) to a cytidine deaminase like APOBEC. Base editors are targeted to a specific locus by a gRNA, and they can convert cytidine to uridine within a small editing window near the PAM site. Uridine is subsequently converted to thymidine through base excision repair, creating a C to T change (or a G to A on the opposite strand).
Likewise, adenine base editors have been engineered to convert adenosine to inosine, which is treated like guanosine by the cell, creating an A to G (or T to C) change. Adenine DNA deaminases do not exist in nature, but have been created by directed evolution of the Escherichia coli TadA, a tRNA adenine deaminase. Like cytosine base editors, the evolved TadA domain is fused to a Cas9 protein to create the adenine base editor.
Both types of base editors are available with multiple Cas9 variants including high fidelity Cas9’s. Further advancements have been made by optimizing expression of the fusions, modifying the linker region between Cas variant and deaminase to adjust the editing window, or adding fusions that increase product purity such as the DNA glycosylase inhibitor (UGI) or the bacteriophage Mu- derived Gam protein (Mu- GAM).
While many base editors are designed to work in a very narrow window proximal to the PAM sequence, some base editing systems create a wide spectrum of single-nucleotide variants (somatic hypermutation) in a wider editing window, and are thus well suited to directed evolution applications. Examples of these base editing systems include targeted AID-mediated mutagenesis (TAM) from the Xing Chang lab and CRISPR-X from the Michael Bassik lab, in which Cas9 is fused to activation-induced cytidine deaminase (AID).
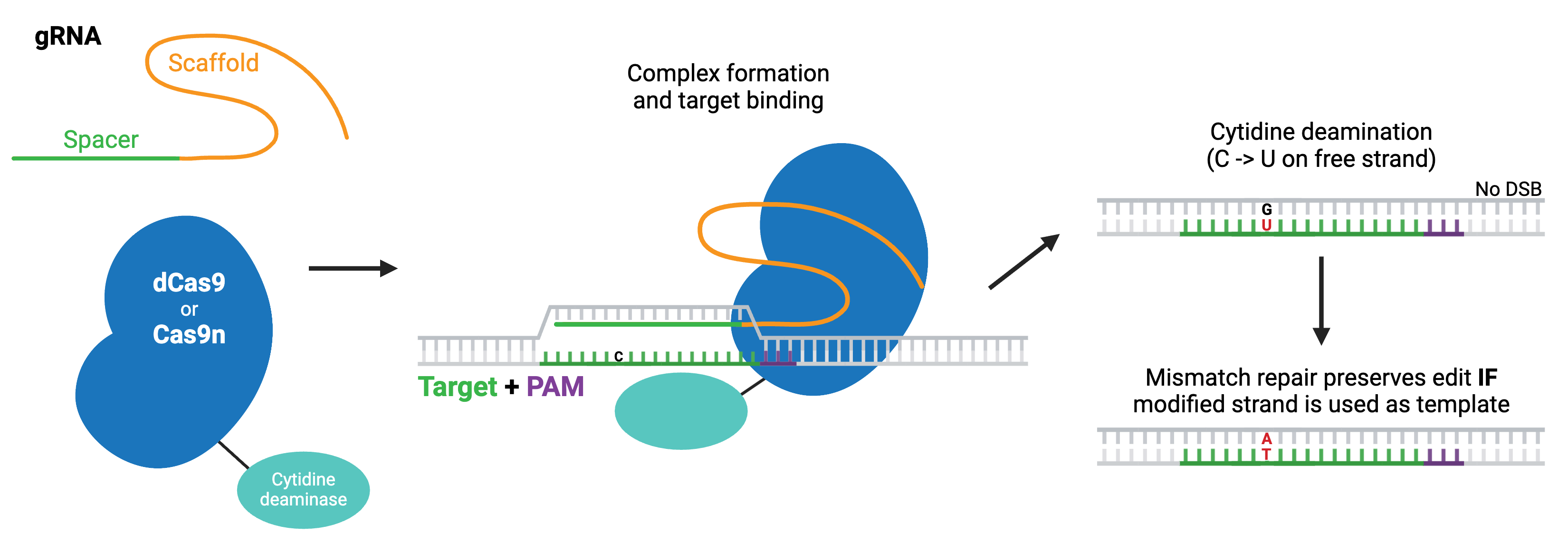
Browse, sort, or search the tables below for CRISPR base editing plasmids. To learn more about base editing and other CRISPR topics, read our CRISPR Guide.
Mammalian
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Bacteria
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Plant
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Yeast
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
Zebrafish
| ID | Plasmid | Gene/Insert | Promoter | Selectable Marker | PI | Publication |
|---|
CRISPR Resources
Addgene has a large selection of CRISPR plasmids and resources. Find more CRISPR functions along with plasmids categorized by organism by visiting our CRISPR plasmids page. Find a comprehensive list of CRISPR resources by visiting our CRISPR reference page.
Content last reviewed: 17 October 2025
Do you have suggestions for other plasmids that should be added to this list?
Fill out our Suggest a Plasmid form or e-mail [email protected] to help us improve this resource!



