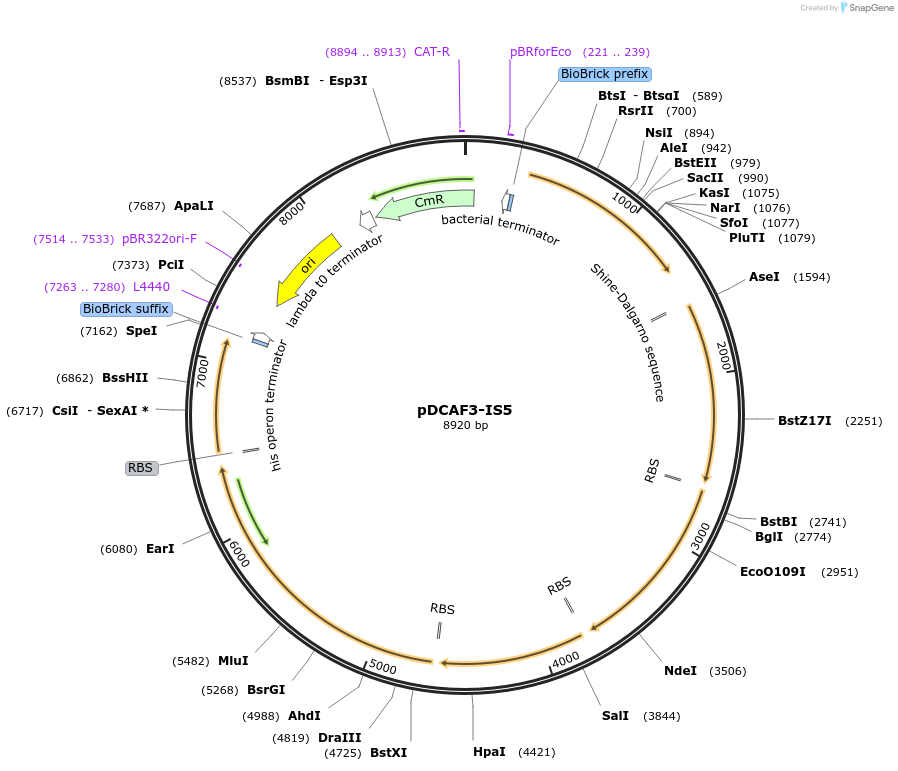
pDCAF3-IS5
(Plasmid
#65220)
-
PurposeExpresses N-demethylases for the degredation of caffeine and related methylxanthines. Can be used to measure caffeine content as described in the publication listed below.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 65220 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 * | |
* Log in to view industry pricing.
Backbone
-
Vector backbonepSB1C3
-
Backbone manufacturerigem parts registry
- Backbone size w/o insert (bp) 2070
- Total vector size (bp) 8920
-
Vector typeBacterial Expression, Synthetic Biology
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol and Kanamycin, 25 & 50 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)BW25113 ΔguaB::kanR
-
Growth instructionsGrow in minimal medium supplemented with 200 uM caffeine, 34 ug/mL chloramphenicol, and 50 ug/mL kanamycin. M9 minimal medium supplemented with 2 g/L glucose and 2 g/L casein (M9GC) is recommended. Plasmid carries chloramphenicol resistance. Strain is auxotrophic for guanine and has a kanamycin resistance marker integrated into the chromosome. Supplementation with caffeine rescues this auxotrophy through the activity of the demethylation enzymes encoded on the plasmid
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert namendmA
-
Alt nameMethylxanthine N1-demethylase
-
SpeciesPseudomonas putida strain CBB5
-
Insert Size (bp)1056
-
GenBank IDJQ061127.1 KC619530.1
- Promoter BBa_J23100
Cloning Information for Gene/Insert 1
- Cloning method Gibson Cloning
- 5′ sequencing primer tgccacctgacgtctaagaa
- 3′ sequencing primer attaccgcctttgagtgagc
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert namendmB
-
Alt nameMethylxanthine N3-demethylase
-
SpeciesPseudomonas putida strain CBB5
-
Insert Size (bp)1068
-
MutationChanged start codon from gtg to atg
-
GenBank IDJQ061128.1 KC619530.1
- Promoter BBa_J23100
Cloning Information for Gene/Insert 2
- Cloning method Gibson Cloning
- 5′ sequencing primer tgccacctgacgtctaagaa
- 3′ sequencing primer attaccgcctttgagtgagc
- (Common Sequencing Primers)
Gene/Insert 3
-
Gene/Insert namendmC
-
Alt nameMethylxanthine N7-demethylase
-
SpeciesPseudomonas putida strain CBB5
-
Insert Size (bp)855
-
GenBank IDJQ061129.1 KC619530.1
- Promoter BBa_J23100
Cloning Information for Gene/Insert 3
- Cloning method Gibson Cloning
- 5′ sequencing primer tgccacctgacgtctaagaa
- 3′ sequencing primer attaccgcctttgagtgagc
- (Common Sequencing Primers)
Gene/Insert 4
-
Gene/Insert namendmD
-
Alt nameMethylxanthine N-demethylase electron transfer protein
-
SpeciesPseudomonas putida strain CBB5
-
Insert Size (bp)1767
-
MutationChanged start codon from gtg to atg.
-
GenBank IDJQ061130.1 KC619530.1
- Promoter BBa_J23100
Cloning Information for Gene/Insert 4
- Cloning method Gibson Cloning
- 5′ sequencing primer tgccacctgacgtctaagaa
- 3′ sequencing primer attaccgcctttgagtgagc
- (Common Sequencing Primers)
Gene/Insert 5
-
Gene/Insert namegst9
-
Alt nameGlutathione S-Transferase.
-
Alt nameHomolog of the ndmE gene from pseudomonas putida CBB5.
-
SpeciesJanthinobacterium sp. Marseille
-
Insert Size (bp)675
-
GenBank IDWP_012081511.1 KC619530.1
- Promoter BBa_J23100
Cloning Information for Gene/Insert 5
- Cloning method Gibson Cloning
- 5′ sequencing primer tgccacctgacgtctaagaa
- 3′ sequencing primer attaccgcctttgagtgagc
- (Common Sequencing Primers)
Gene/Insert 6
-
Gene/Insert namechlR
-
Alt nameCAT. Chloramphenicol acetyltransferase.
-
Insert Size (bp)660
-
GenBank IDKC619530.1
Resource Information
-
Supplemental Documents
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
The sequence of this plasmid is updated from the GenBank entry for pDCAF3 (KC619530.1) to include an IS5 insertion that occurred in the promoter region of the caffeine demethylation genes during passaging of the strain and a base change in the sequence of the gst9 gene. This plasmid still functions as described in the publication for pDCAF3.
Vector constitutively expresses N-demethylase genes under the control of the BBa_J23100 promoter (http://parts.igem.org/Part:BBa_J23100)
N-demethylases are only functional when grown at 30°C and require iron supplementation in minimal media. See:
Summers, R. M., Louie, T. M., Yu, C.-L., Gakhar, L., Louie, K.C., and Subramanian, M. (2012) Novel, highly specific Ndemethylases enable bacteria to live on caffeine and related purine alkaloids. J. Bacteriol. 194, 2041−2049.
For the quantification of caffeine (or other methylxanthine) content it is recommended to grow cells in M9 minimal medium supplemented with 2 g/L glucose and 2 g/L casein (M9GC).
Please note that this plasmid may require a unique bacterial strain, so make sure to confirm that you can also obtain the appropriate growth strain. Please contact us at [email protected] or contact our distributors if you have any questions.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pDCAF3-IS5 was a gift from Jeffrey Barrick (Addgene plasmid # 65220 ; http://n2t.net/addgene:65220 ; RRID:Addgene_65220) -
For your References section:
Decaffeination and measurement of caffeine content by addicted Escherichia coli with a refactored N-demethylation operon from Pseudomonas putida CBB5. Quandt EM, Hammerling MJ, Summers RM, Otoupal PB, Slater B, Alnahhas RN, Dasgupta A, Bachman JL, Subramanian MV, Barrick JE. ACS Synth Biol. 2013 Jun 21;2(6):301-7. doi: 10.1021/sb4000146. Epub 2013 Mar 22. 10.1021/sb4000146 PubMed 23654268