-
Depositing Lab
-
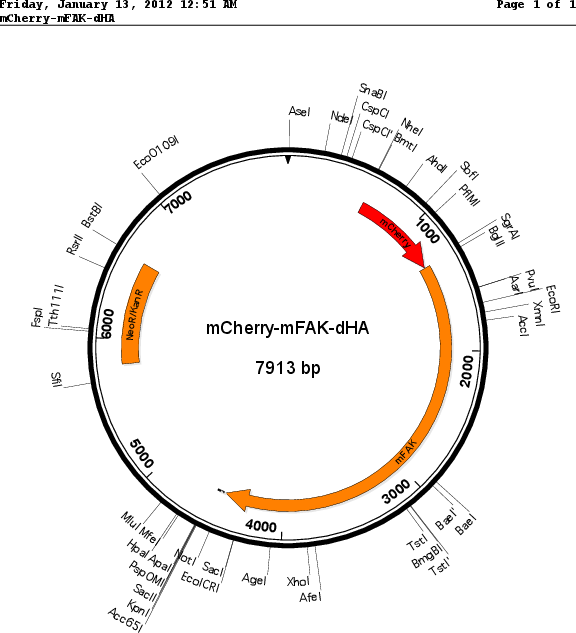
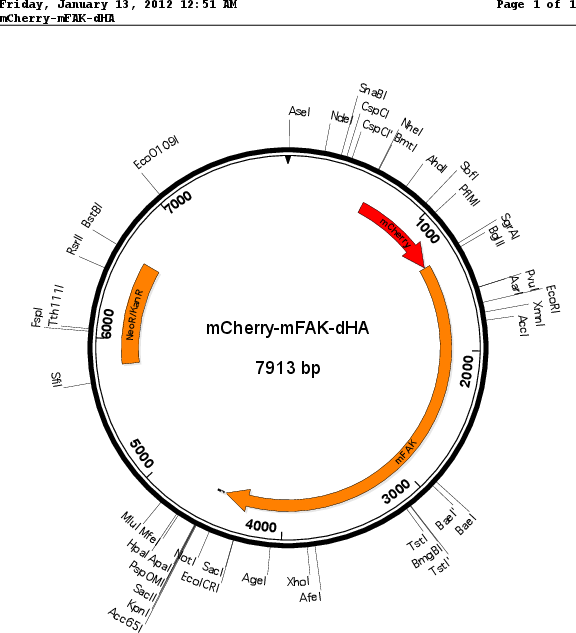
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 35039 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepmCherry-c1
-
Backbone manufacturerBD Clontech
- Backbone size w/o insert (bp) 4700
-
Vector typeMammalian Expression
-
Selectable markersNeomycin (select with G418)
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert namefocal adhesion kinase
-
Alt nameFAK
-
SpeciesM. musculus (mouse)
-
Insert Size (bp)3150
-
MutationY42H; deletion of amino acids 1051 & 1052
-
Entrez GenePtk2 (a.k.a. FADK 1, FAK, FRNK, Fadk, p125FAK)
-
Tags
/ Fusion Proteins
- mCherry (N terminal on insert)
- 2x HA (C terminal on insert)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site Bgl 2 (not destroyed)
- 3′ cloning site Kpn 1 (not destroyed)
- 5′ sequencing primer GACAGATCTATGGCAGCTGCTTATCTTGACCCAAAC
- 3′ sequencing primer 5' GTAGGTACCTTATCTAGATCCGGTGGATC 3' (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byMurine GFP-FAK (pEGFP-C1-FAK-HA) was provided by D. Schlaepfer (University of California San Diego).
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please note that Addgene's sequencing result identified a Y42H mutation when compared to the full plasmid sequence provided by the depositing laboratory and to GenBank entry NP_032008.2.
The final two FAK amino acids are missing in this construct, as the original source for the FAK construct used in this plasmid was originally missing these same amino acids. The depositing laboratory states that the deletion should not affect any FAK function since it is outside of the FAK primary sequence.
Addgene's sequencing results also identified a deletion region from bp#4553-4588 when compared to the full plasmid sequence provided by the depositing laboratory. This indicates that the restriction sites KpnI (4561), SacII (4564) and ApaI (4569) are not present in the vector.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pmCherry-C1-FAK-HA was a gift from Anna Huttenlocher (Addgene plasmid # 35039 ; http://n2t.net/addgene:35039 ; RRID:Addgene_35039) -
For your References section:
Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). Chan KT, Bennin DA, Huttenlocher A. J Biol Chem. 2010 Apr 9;285(15):11418-26. Epub 2010 Feb 11. 10.1074/jbc.M109.090746 PubMed 20150423