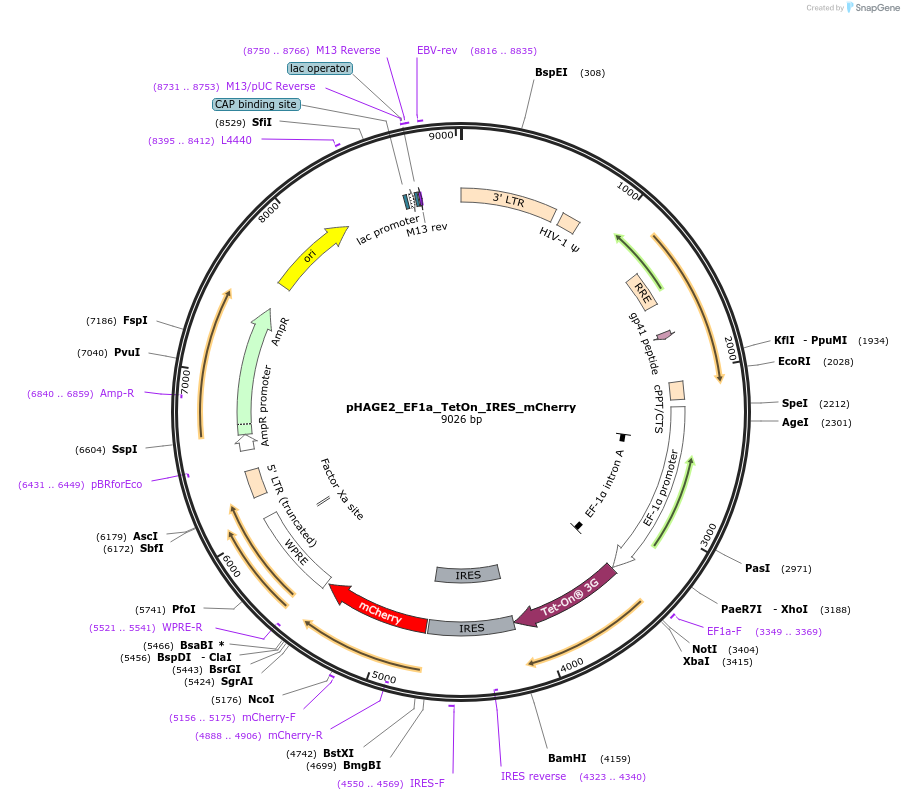
-
PurposeLentivirus backbone expressing rtTA linked to mCherry
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 204155 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepHAGE2
- Total vector size (bp) 9026
-
Vector typeMammalian Expression, Lentiviral
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameTet-On 3G
-
Insert Size (bp)745
- Promoter EF1a
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site NotI (unknown if destroyed)
- 3′ cloning site BamHI (not destroyed)
- 5′ sequencing primer agcctcagacagtggttcaaagt
- 3′ sequencing primer ccttattccaagcggcttcgg (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pHAGE2_EF1a_TetOn_IRES_mCherry was a gift from Jesse Bloom (Addgene plasmid # 204155 ; http://n2t.net/addgene:204155 ; RRID:Addgene_204155) -
For your References section:
A pseudovirus system enables deep mutational scanning of the full SARS-CoV-2 spike. Dadonaite B, Crawford KHD, Radford CE, Farrell AG, Yu TC, Hannon WW, Zhou P, Andrabi R, Burton DR, Liu L, Ho DD, Chu HY, Neher RA, Bloom JD. Cell. 2023 Mar 16;186(6):1263-1278.e20. doi: 10.1016/j.cell.2023.02.001. Epub 2023 Feb 13. 10.1016/j.cell.2023.02.001 PubMed 36868218