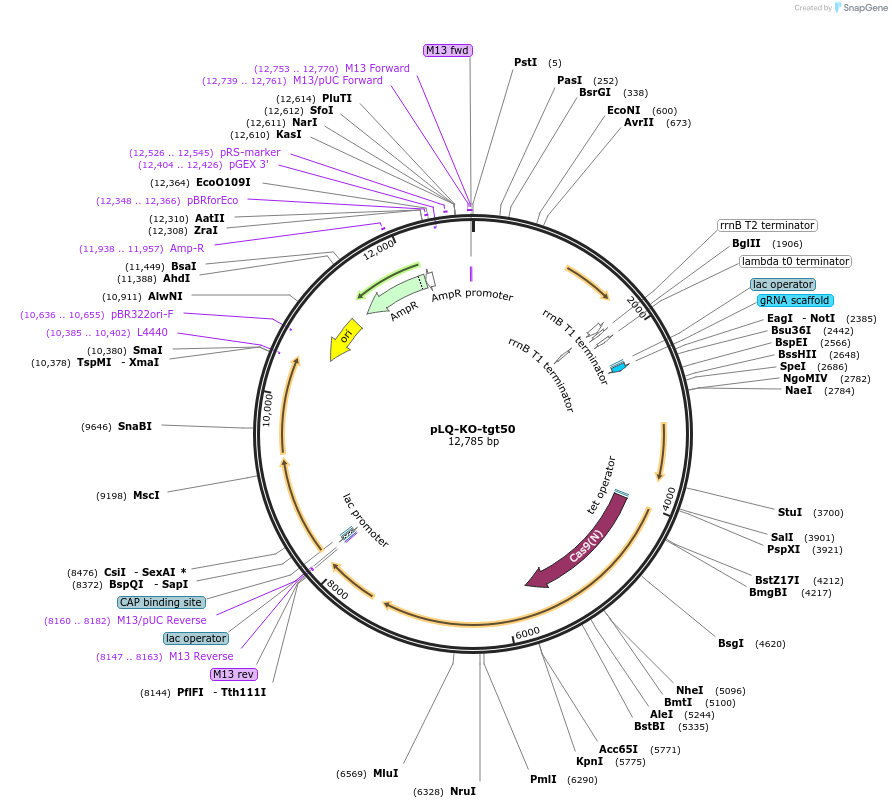
pLQ-KO-tgt50
(Plasmid
#126578)
-
PurposeCRISPR-Cas9 based efficient genome editing in S. aureus
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 126578 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backboneColE1 origin, AmpR, Rep(+), cat resistance
- Backbone size w/o insert (bp) 4000
-
Vector typeBacterial Expression, CRISPR
-
Selectable markersChloramphenicol in S. aureus.
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Growth instructionsHigh copy in E. coli, low copy in S. aureus. chloramphenicol resistance in S. aureus
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameCas9-Pxyltet-sgRNA-Pspac-donor-tgt50
-
SpeciesS. pyogenes
-
Insert Size (bp)8000
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Addgene NGS identified a D24E and C-terminal truncation of Cas9. The depositing lab has confirmed that these should not affect function.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pLQ-KO-tgt50 was a gift from Sheng Yang (Addgene plasmid # 126578 ; http://n2t.net/addgene:126578 ; RRID:Addgene_126578) -
For your References section:
CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Liu Q, Jiang Y, Shao L, Yang P, Sun B, Yang S, Chen D. Acta Biochim Biophys Sin (Shanghai). 2017 Sep 1;49(9):764-770. doi: 10.1093/abbs/gmx074. 10.1093/abbs/gmx074 PubMed 28910979