-
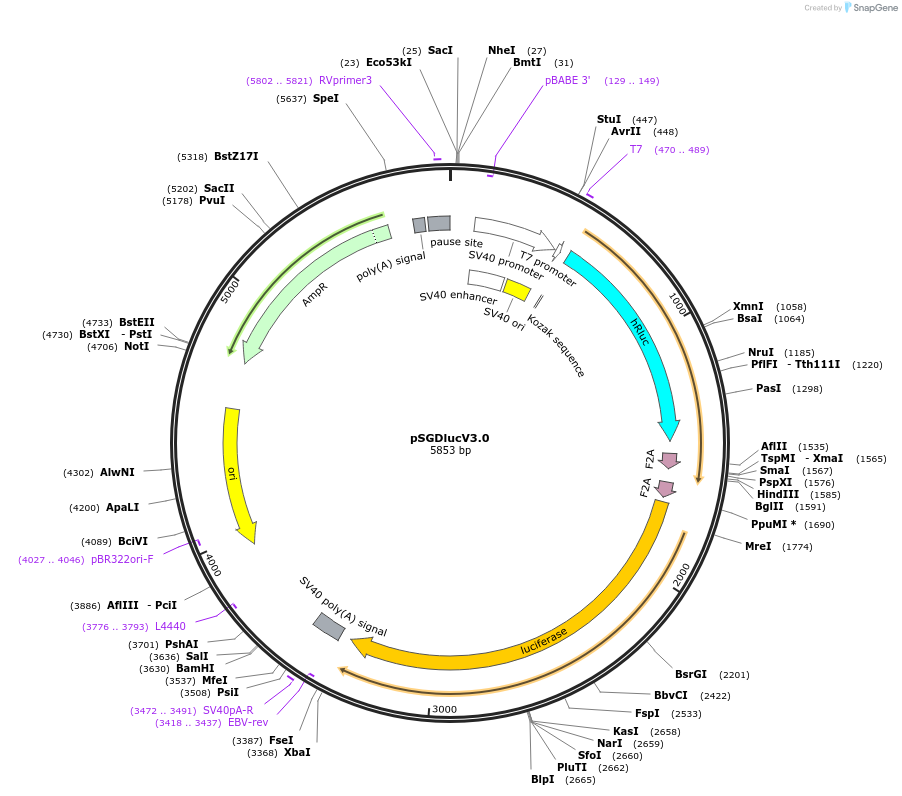
Purpose(Empty Backbone) dual luciferase with FMDV StopGo sequences flanking MCS
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 119760 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepBR322
- Backbone size (bp) 5853
-
Vector typeMammalian Expression, Luciferase
- Promoter SV40 and T7
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Cloning Information
- Cloning method Restriction Enzyme
- 5′ sequencing primer T7 (Common Sequencing Primers)
Resource Information
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
This plasmid is an updated version of the original pSGDluc (https://www.addgene.org/87323/)
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pSGDlucV3.0 was a gift from John Atkins (Addgene plasmid # 119760 ; http://n2t.net/addgene:119760 ; RRID:Addgene_119760) -
For your References section:
Lack of evidence for ribosomal frameshifting in ATP7B mRNA decoding. Loughran G, Fedorova AD, Khan YA, Atkins JF, Baranov PV. Mol Cell. 2022 Oct 6;82(19):3745-3749.e2. doi: 10.1016/j.molcel.2022.08.024. Epub 2022 Sep 16. 10.1016/j.molcel.2022.08.024 PubMed 36115342