We narrowed to 5,132 results for: codon optimized
-
Plasmid#171687PurposepRS316-PTEF1-CnoCas9, contains Cryptococcus neoformans codon optimized Cas9 under the control of the TEF1 promoter and terminator.DepositorInsertCnoCAS9
UseCRISPRExpressionBacterial and YeastPromoterTEF1Available SinceMay 4, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRiboI-APEX2m
Plasmid#176843PurposeExpression of APEX2 from a codon-optimized gene (single-copy integrating plasmid)DepositorInsertpRibo-APEX2m
UseIntegratingExpressionBacterialPromoterpRibo, theophilline riboswitch inducible hsp60 pr…Available SinceJan. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
pRibo-APEX2m
Plasmid#176842PurposeExpression of APEX2 from a codon-optimized gene (multi-copy episomal plasmid)DepositorInsertpRibo-APEX2m
ExpressionBacterialPromoterpRibo, theophilline riboswitch inducible hsp60 pr…Available SinceJan. 10, 2022AvailabilityAcademic Institutions and Nonprofits only -
pmCellFree_KA1_SARS-COV-2_ORF7a
Plasmid#169393PurposeCell-free/mammalian expression vector for SARS-COV-2 virus ORF7a with N-term eGFPDepositorInsertSARS-COV-2 _ORF7a (ORF7a SARS-Cov2)
UseCell-free expression vectorTagsHis and eGFPMutationCodon optimized for Saccharomyces cerevisiaePromoterT7- CMV/CAGAvailable SinceJuly 8, 2021AvailabilityAcademic Institutions and Nonprofits only -
pML597
Plasmid#32235PurposeExpresses M. tuberculosis codon-optimized flpe geneDepositorInsertmflpe
ExpressionBacterialMutationM.tuberculosis codon optimized flpe geneAvailable SinceNov. 18, 2011AvailabilityAcademic Institutions and Nonprofits only -
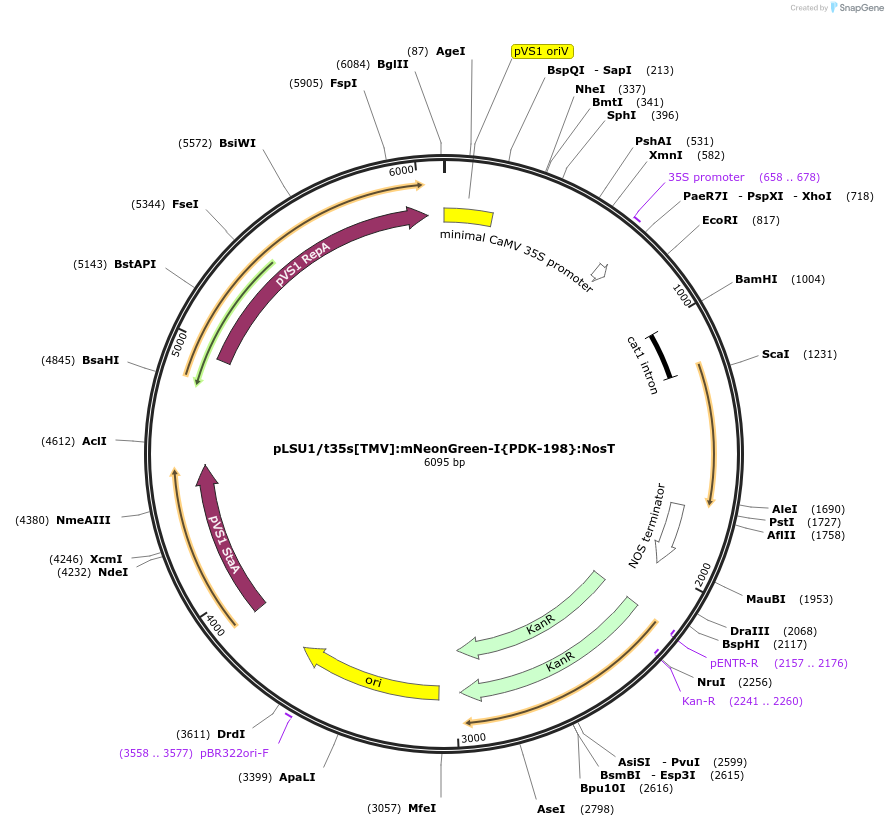
pLSU1/t35s[TMV]:mNeonGreen-I{PDK-198}:NosT
Plasmid#212170PurposeThis binary vector expresses mNeonGreen containing the castor bean catalase (PDK) intron (198bp into the gene) with the truncated 35sCAMV promoter and the tomato mosaic virus UTRDepositorInsertTomato codon optimized version of mNeonGreen-I{PDK-198}
ExpressionPlantMutationmNeonGreen codon-optimized for tomato and PDK int…Promotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
pLSU1/t35s[TMV]:mNeonGreen-I{AGO-198}:NosT
Plasmid#212169PurposeThis binary vector expresses mNeonGreen containing the Argonaut (AGO) intron (198bp into the gene) with the truncated 35sCAMV promoter and the tomato mosaic virus UTRDepositorInsertTomato codon optimized version of mNeonGreen-I{AGO-198}
ExpressionPlantMutationmNeonGreen codon-optimized for tomato and AGO int…Promotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
pTEF:ATP
Plasmid#92179PurposeHO-TEF1p-ATeam1.03-KanMX4-HO. Codon optimized for yeast version of the ATeam1.03 ATP FRET sensor (originally from Imamura et al. PNAS, 2009 ), expressed via the TEF1 promoter. HO locus integration.DepositorInsertATeam1.03 codon optimized for yeast (Saccharomyces cerevisiae)
UseIntegration and expression into yeast.TagsYeast codon optimized mseCFP (donor) and cp173-mV…ExpressionYeastPromoterTEF1pAvailable SinceJune 26, 2017AvailabilityAcademic Institutions and Nonprofits only -
pAaePUb::Cas9-2A-Neo
Plasmid#176680PurposeExpression of Drosophila codon optimized spCas9 under Ae. aegypti Polyubiquitin promoter (AAEL003888)DepositorInsertspCas9; NeoR (aminoglycoside phosphotransferase from Tn5)
UseCrisprExpressionInsectMutationDrosophila codon optimizedPromoterAe. aegypti poly-ubiquitin (AAEL003888)Available SinceNov. 24, 2021AvailabilityAcademic Institutions and Nonprofits only -
pSF11-wNIR-GECO2-T2A-HO1
Plasmid#159606PurposeCodon-optimized near-infrared fluorescent calcium indicator NIR-GECO2 with codon-optimized heme-oxygenase in C.elegans expression plasmidDepositorInsertwNIR-GECO2-T2A-HO1
ExpressionWormPromotertag-168Available SinceSept. 22, 2020AvailabilityAcademic Institutions and Nonprofits only -
pTD-C_eYFPTwinStrep
Plasmid#45942Purposeexpresses E.coli codon optimized EYFP-Twin-Strep fusion proteinDepositorInsertsynthetic eYFP (codon usage adapted to E.coli K12)
TagsTwin-Strep-tagExpressionBacterialPromoterlacIq/PtrcAvailable SinceSept. 18, 2013AvailabilityAcademic Institutions and Nonprofits only -
Dm_λHA-FLuc100_CSC
Plasmid#210592PurposeExpresses lambdaHA-optimal Firefly (CSC metric; Drosophila)DepositorInsertLambdaHA-optimal Firefly luciferase (Dm optimization, CSC metric)
Tagslambda HAExpressionInsectPromoterAc5Available SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pUB_SMFLuc100---MS2
Plasmid#210570PurposeExpresses SM(FLAG)-optimal Firefly with 3'UTR MS2 hairpinsDepositorInsertSpaghetti monster (FLAG)-optimal FLuc
UseLentiviral and LuciferaseTagsSpaghetti monster (FLAG)ExpressionMammalianPromoterUBC humanAvailable SinceOct. 23, 2024AvailabilityAcademic Institutions and Nonprofits only -
pLSU1/t35s[]:mNeonGreen-CI{PIV-198}:NosT
Plasmid#212171PurposeThis binary vector expresses mNeonGreen containing the PIV intron (198bp into the gene) with extra 3' splice site removed (corrected intron) with the truncated 35sCAMV promoterDepositorInsertTomato codon optimized version of mNeonGreen-I{PIV-198}
ExpressionPlantMutationmNeonGreen codon-optimized for tomato and PIV int…Promotertruncated 35sCaMVAvailable SinceMarch 6, 2024AvailabilityAcademic Institutions and Nonprofits only -
JBEI-3716
Plasmid#100169PurposeUsed to improve production from engineered biosynthetic pathways include optimizing codon usage, enhancing production of rate-limiting enzymes, and eliminating the accumulation of toxic intermediates or byproducts to improve cell growthDepositorInsertcodon-optimized MK
ExpressionBacterialAvailable SinceSept. 15, 2017AvailabilityAcademic Institutions and Nonprofits only -
pSM-mStayGold
Plasmid#234317Purposecodon-optimized mStayGold with synthetic introns for the expression in C. elegansDepositorInsertC. elegans codon optimized mStayGOld
TagsmStayGoldExpressionWormAvailable SinceMarch 26, 2025AvailabilityAcademic Institutions and Nonprofits only -
pFastBac_Dual_SNF2L-opt_p48
Plasmid#102243PurposeExpression of human codon optimized human SNF2L and pRbAp48 from pFastBac dualDepositorExpressionInsectMutationcodon optimized for expressionPromoterPolyhedrin and p10Available SinceNov. 1, 2017AvailabilityIndustry, Academic Institutions, and Nonprofits -
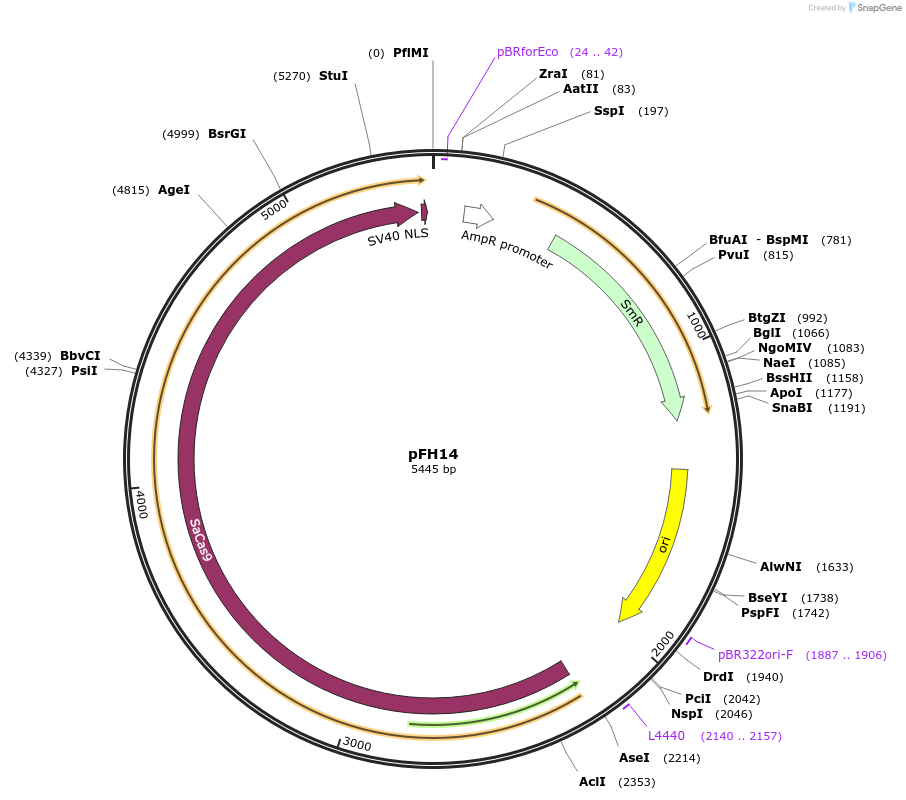
pFH14
Plasmid#125799PurposeLevel0 SaCas9, Arabidopsis codon optimizedDepositorInsertSaCas9, Arabidopsis codon optimized
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pFH47
Plasmid#125812PurposeLevel0 LbCas12a, human codon optimizedDepositorInsertLbCas12a, human codon optimized
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only -
pFH13
Plasmid#125798PurposeLevel0 SpCas9, wheat codon optimizedDepositorInsertSpCas9, wheat codon optimized
UseCRISPR and Synthetic BiologyExpressionPlantAvailable SinceAug. 19, 2019AvailabilityAcademic Institutions and Nonprofits only