Plasmid Pooled Libraries
Pooled libraries represent a powerful tool for forward genetic screening, or identifying previously unknown genes that contribute to a phenotype. At their simplest level, pooled libraries are single preparations of many different plasmids. Plasmids within a given library have the same backbone, but they express or target different genes. Some libraries cover the majority of the genome, while others are restricted to certain gene sets. In cDNA libraries, each plasmid contains a unique cDNA. In shRNA or gRNA libraries, each plasmid contains a unique gene targeting sequence, but there are multiple sequences targeting each gene in the overall library. Barcoding libraries contain plasmids with unique, semi-random sequences that can be used for applications like lineage tracing or parsing the effects of expressing multiple genes at once. A well-designed screen can help you begin to understand what genes are important to a certain phenotype, and allow you to design additional hypothesis-directed experiments.
As you might have guessed from their name, pooled libraries are supplied as a mixed population of plasmids in a single tube. Pooled libraries can be small, if they are designed to cover only a subset of genes, or very large (e.g., the Toronto KnockOut (TKO) library from the Moffat lab has over 175,000 different gRNA-containing plasmids).
Amplifying and Using the Library
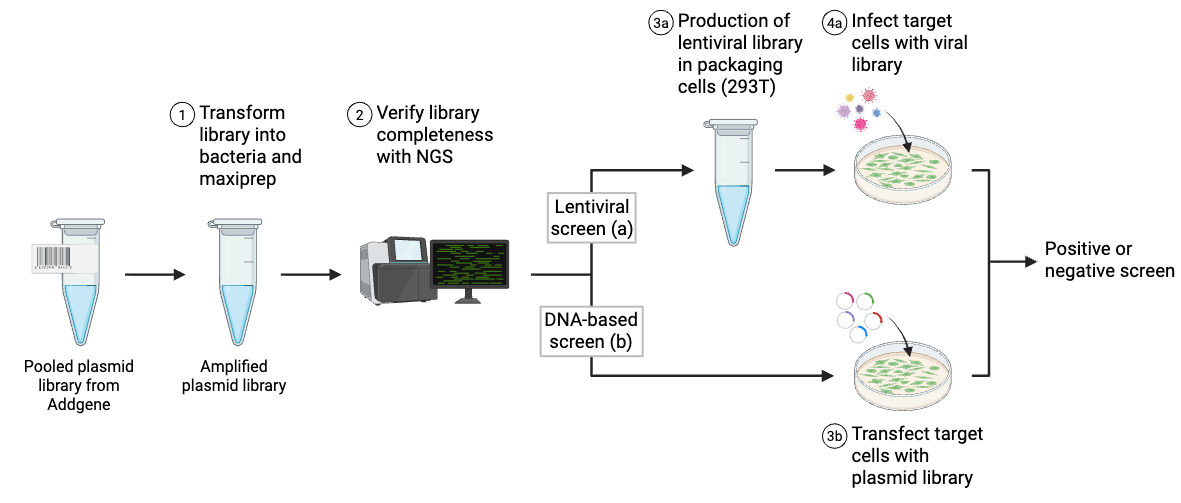
Once you receive your library from Addgene, check the library information to see if it should be amplified before you conduct your screen. If you need to amplify the library, please refer to the depositor’s protocol for the best results.
For some libraries, plasmid DNA can be delivered directly to the cells of interest. With others, notably pooled lentiviral plasmid libraries, the plasmids must first be used to make virus. This pooled virus is subsequently used to deliver the plasmids to the cells of interest. In either case, next-generation sequencing of the maxiprep DNA is recommended to verify that the library is complete — an incomplete library may lead to false positives or false negatives in later experiments, and can also negatively affect data reproducibility.
Types of Pooled Library Screens
In pooled library screens, cells are infected at a very low multiplicity of infection (MOI) to increase the odds that an infected cell receives only one plasmid. To make sure that every plasmid is adequately represented in the population, you’ll need to infect many more cells than the number of plasmids present in the library — for example, the Moffat lab uses 200x the number of cells as plasmids in the TKO library. Using large numbers of cells minimizes the effects of random chance that could lead to false positive or negative results — for a given plasmid, think of each cell carrying it as being a biological replicate for that plasmid.
Library screens can be divided into two types: positive screens and negative screens (Figure 2). Both types of screen employ a selection method relevant to the phenotype being studied. Examples of published selection mechanisms for lentiviral CRISPR libraries include: resistance to the anti-cancer drug vemurafenib , resistance to Clostridium septicum alpha-toxin , and resistance to to the nucleotide analog 6-thioguanine . Next-generation sequencing is the only way to accurately evaluate which library components are influenced by selection, so it’s essential that you have access to this technology if you’re planning to conduct a pooled library screen.
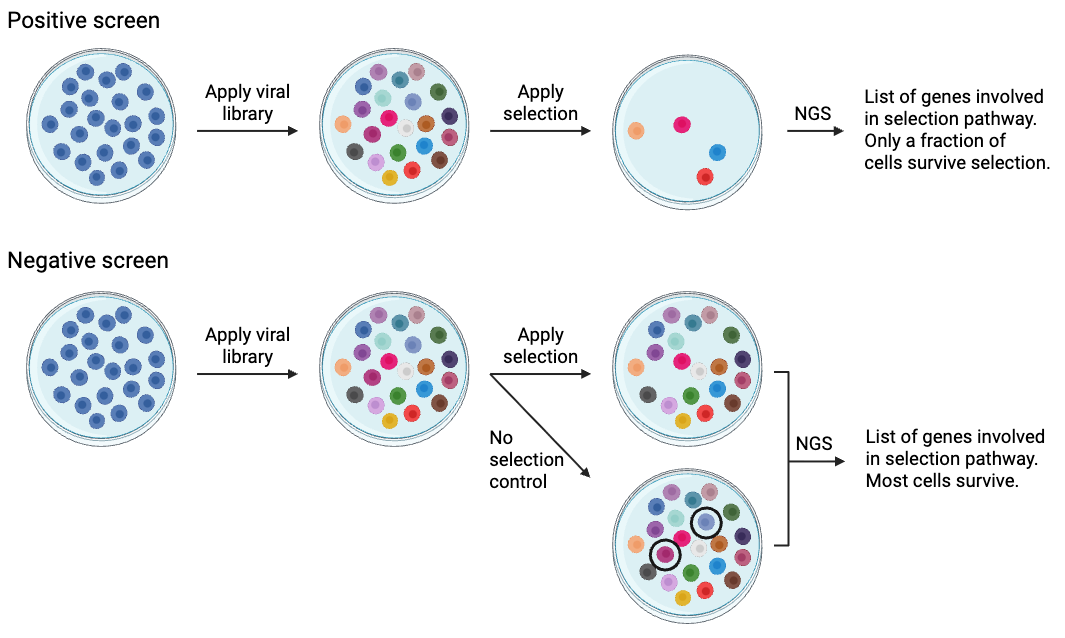
Both positive and negative screens follow the same basic process:
- Amplify the library (electroporation and maxiprep).
- If delivering as virus, make virus; this creates a pooled lentiviral CRISPR library.
- Apply pooled library to cells.
The screens diverge at a few important points, including when to apply selection, when to sequence, and how to analyze the results:
POSITIVE screen:
- Apply library
- Apply selection
- Most cells die or do not pass selection (in the case of a reporter)
- Sort “winning” cells
- Sequence those cells that pass (“win”)
- Get a list of genes involved in the selection
NEGATIVE screen:
- Apply library
- Perform Next Generation Sequencing (NGS) on a control sample (no selection)
- In parallel, apply library
- Apply selection; most cells will survive
- Perform NGS on the surviving cells
- Compare NGS results between experimental and control cells
- Generate a list of gRNAs that disappear with addition of the selection mechanism
Positive Screen
In a positive screen, the goal is to identify those cells that survive post-selection. The selective pressure must be strong enough that most of the cells die, removing their plasmids from the population, and only a small fraction survive. After the surviving cells are collected, their plasmids are PCR-amplified and sequenced using NGS to identify their cDNA or target gene. Positive selection screens are generally very robust, and tens of thousands of genes that were initially present may be narrowed down to a few hundred or fewer ‘winners’.
Negative Screen
Negative screens are a little trickier than positive screens. In a negative screen, the goal is to identify those cells that do not survive the selection mechanism. You’ll infect two sets of cells and subject one set to selection while the other serves as a non-selected control. These two populations are then sequenced using NGS to determine which plasmids have been depleted by selection. In a CRISPR screen, negative screens are often used to identify genes that are essential for growth/survival under certain conditions.
Next-Generation Sequencing
Next-generation sequencing allows for parallel sequencing of millions of DNA fragments simultaneously in a single reaction. As discussed above, access to this technology is crucial for conducting pooled library screens. Addgene does not offer next-generation sequencing services. For an NGS company near you, please consult these links:
NGS Company Resources
Resources
Find additional resources on our website:
- See our Pooled Library Amplification Protocol for Addgene's generalized library amplification protocol.
- Read our CRISPR guide for more information on genome-wide screening with CRISPR/Cas9.
- Browse our Viral Vector resources.
- Check out our blog posts featuring some of our most popular libraries!


