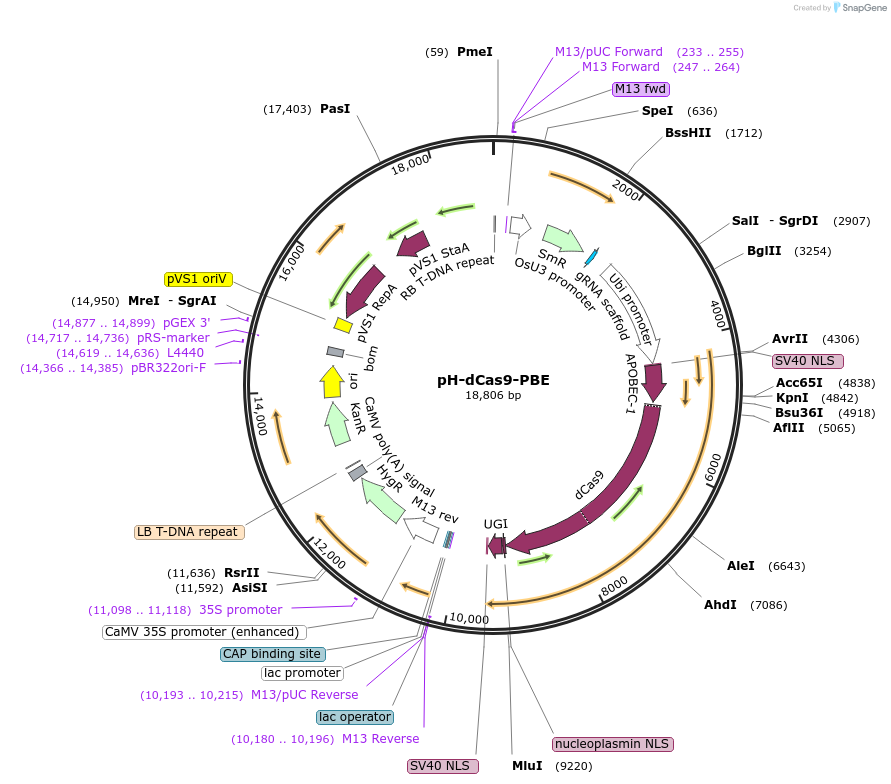
-
PurposeExpresses CRISPR base editor in rice
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 98162 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepHUE411
-
Vector typePlant Expression ; Rice
-
Selectable markersHygromycin
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameNLS-APOBEC1-XTEN-dCas9-UGI-NLS
- Promoter Ubi
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer Unknown
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byAPOBEC1, XTEN, dCas9 (D10A and H840A) and UGI sequences were codon-optimized for cereal plants, and synthesized commercially (GenScript, Nanjing, China).
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pH-dCas9-PBE was a gift from Caixia Gao (Addgene plasmid # 98162 ; http://n2t.net/addgene:98162 ; RRID:Addgene_98162) -
For your References section:
Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Nat Biotechnol. 2017 May;35(5):438-440. doi: 10.1038/nbt.3811. Epub 2017 Feb 27. 10.1038/nbt.3811 PubMed 28244994