-
Purpose(Empty Backbone) Doyon lab Tandem-Affinity Purification Following Nuclease-Driven Gene Addition to the AAVS1 Genomic Safe Harbor Locus. Transgene expression is controlled by an Auto-Regulated Tet-On 3G System.
-
Depositing Lab
-
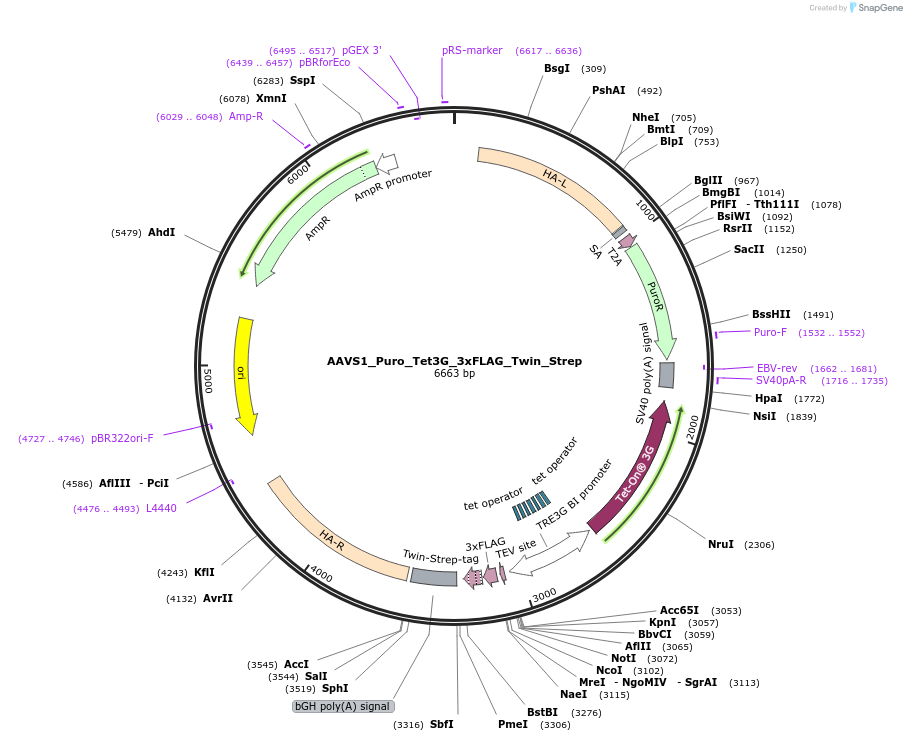
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 92099 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepUC19
- Backbone size (bp) 2364
-
Modifications to backboneInsert was cloned into PvuII digested pUC19 (loss of 322bp including MCS)
-
Vector typeMammalian Expression, CRISPR, TALEN ; ZFN
- Promoter Tet-On 3G Bidirectional
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer See protocol
- 3′ sequencing primer See protocol
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byModified from AAVS1 SA-2A-puro-pA donor (Plasmid #22075). New backbone, overall structure is similar but several modifications have been made through gene synthesis.
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
A detailed protocol for purification of protein complexes tagged with this vector can be found in Doyon & Côté (2016). Preparation and Analysis of Native Chromatin-Modifying Complexes. Methods in Enzymology, 573, 303–318. https://doi.org/10.1016/bs.mie.2016.01.017. https://www.ncbi.nlm.nih.gov/pubmed/27372759
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
AAVS1_Puro_Tet3G_3xFLAG_Twin_Strep was a gift from Yannick Doyon (Addgene plasmid # 92099 ; http://n2t.net/addgene:92099 ; RRID:Addgene_92099) -
For your References section:
A Scalable Genome-Editing-Based Approach for Mapping Multiprotein Complexes in Human Cells. Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Cote J, Doyon Y. Cell Rep. 2015 Oct 7. pii: S2211-1247(15)01020-7. doi: 10.1016/j.celrep.2015.09.009. 10.1016/j.celrep.2015.09.009 PubMed 26456817