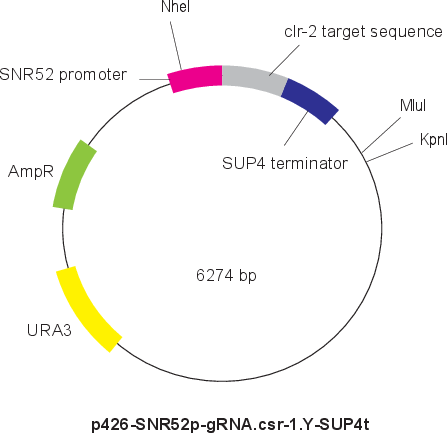
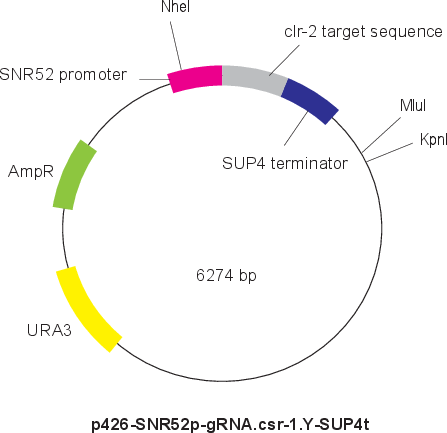
p426-SNR52p-gRNA.clr-2.Y-SUP4t
(Plasmid
#89692)
-
PurposegRNA for clr-2 locus
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 89692 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepRS426
-
Backbone manufacturerATCC
- Backbone size w/o insert (bp) 5726
- Total vector size (bp) 6274
-
Modifications to backboneThe target sequence to csr-1 locus was added instead of CAN1 of p426-SNR52p-gRNA.CAN1.Y-SUP4t (Plasmid #43803) from Dr. Church lab.
-
Vector typeCRISPR ; gRNA
-
Selectable markersURA3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert namegRNA for clr-2 locus
-
Alt namegRNA
-
Alt nameclr-2
-
gRNA/shRNA sequenceGAGTGCCCTAGTCGGTGTGA
-
SpeciesN. crassa
-
Tag
/ Fusion Protein
- No
Cloning Information
- Cloning method Ligation Independent Cloning
- 5′ sequencing primer GGAGCTTCTTTGAAAAGATAATGTATG
- 3′ sequencing primer GTTACATGCGTACACGCGTC (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
p426-SNR52p-gRNA.clr-2.Y-SUP4t was a gift from Christian Hong (Addgene plasmid # 89692 ; http://n2t.net/addgene:89692 ; RRID:Addgene_89692) -
For your References section:
Efficient gene editing in Neurospora crassa with CRISPR technology. Matsu-Ura T, Baek M, Kwon J, Hong C. Fungal Biol Biotechnol. 2015 Jul 15;2:4. doi: 10.1186/s40694-015-0015-1. eCollection 2015. 10.1186/s40694-015-0015-1 PubMed 28955455