pLacZa-GFP
(Plasmid
#86440)
-
Purposevisualization and quantifying the out-of-frame mutations from genome editing
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 86440 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
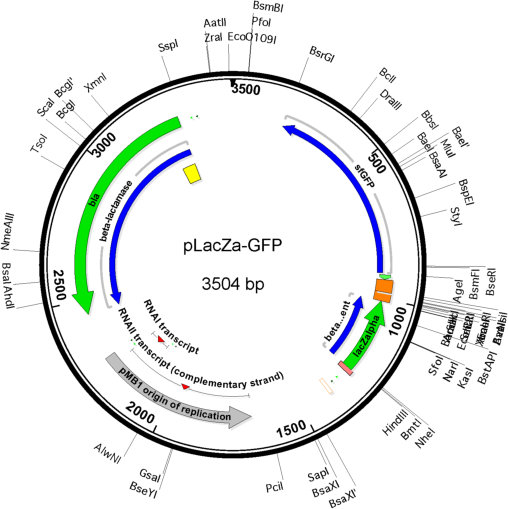
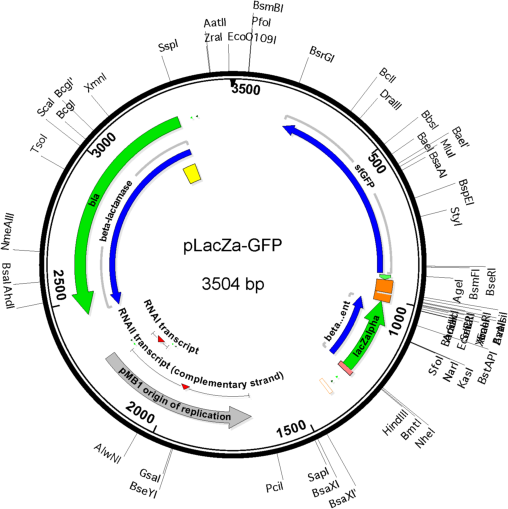
Vector backbonepUC19
-
Backbone manufacturerThermofischer
- Backbone size w/o insert (bp) 2687
- Total vector size (bp) 3504
-
Vector typeColor visualization in bacteria
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameLacZalpha-MCS-sfGFP fusion gene
-
Insert Size (bp)1072
- Promoter Plac
-
Tag
/ Fusion Protein
- LacZa-sfGFP
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site HindIII (not destroyed)
- 3′ cloning site NdeI (destroyed during cloning)
- 5′ sequencing primer M13R (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byLacZa was PCR amplified from pUC19 of Thermofischer and sfGFP gene was synthesized based on sequences of GenBank # HQ873313. sfGFP gene was synthesized based on sequences of GenBank # HQ873313.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
The whole plasmid sequences were compiled based on vector sequences information. It was not confirmed by DNA sequencing.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pLacZa-GFP was a gift from Yun-Bo Shi (Addgene plasmid # 86440 ; http://n2t.net/addgene:86440 ; RRID:Addgene_86440) -
For your References section:
A simple and efficient method to visualize and quantify the efficiency of chromosomal mutations from genome editing. Fu L, Wen L, Luu N, Shi YB. Sci Rep. 2016 Oct 17;6:35488. doi: 10.1038/srep35488. 10.1038/srep35488 PubMed 27748423