-
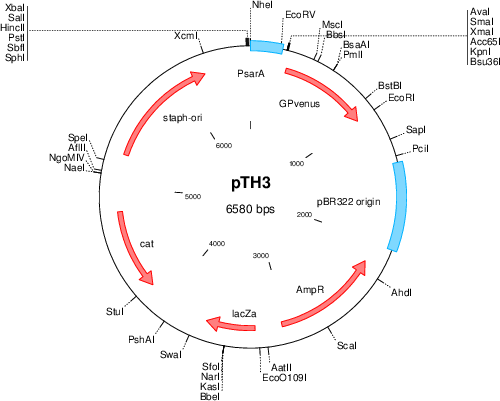
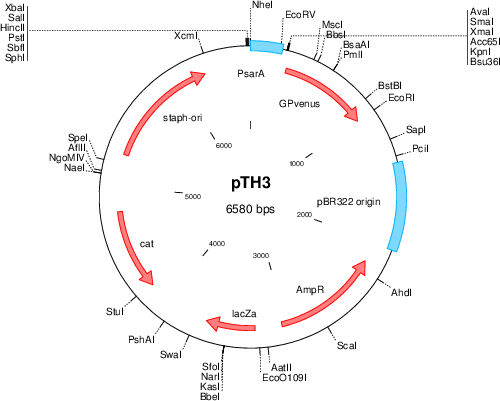
Purposeshuttle plasmid for sarA P1-YFP expression
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 84453 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepCM29
- Backbone size w/o insert (bp) 6604
- Total vector size (bp) 6580
-
Modifications to backbonesGFP replaced with insert
-
Vector typeshuttle vector E.coli-S.aureus, expression of sGFP from the SarA-P1 promoter
-
Selectable markersalso contains Chloramphenicol resistance gene, but only functional in Gram-postive bacteria.
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)Top10F
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameYFP venus
-
Insert Size (bp)737
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site kpnI (not destroyed)
- 3′ cloning site EcoRI (not destroyed)
- 5′ sequencing primer TTGCATGCCTGCAGGTCGACTCTA
- 3′ sequencing primer TTATGCTTCCGGCTCGTATGTTGTGTGG (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byYFP / Venus amplified from pJL77; Liese J, Rooijakkers SHM, van Strijp JAG, Novick RP, Dustin ML. 2013. Intravital two-photon microscopy of host–pathogen interactions in a mouse model of Staphylococcus aureus skin abscess formation. Cellular microbiology 15:891.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pTH3 was a gift from Reindert Nijland (Addgene plasmid # 84453 ; http://n2t.net/addgene:84453 ; RRID:Addgene_84453) -
For your References section:
Fluorescent reporters for markerless genomic integration in Staphylococcus aureus. de Jong NW, van der Horst T, van Strijp JA, Nijland R. Sci Rep. 2017 Mar 7;7:43889. doi: 10.1038/srep43889. 10.1038/srep43889 PubMed 28266573