-
PurposeExpression of mCherry tagged BUB3 in human cells
-
Depositing Lab
-
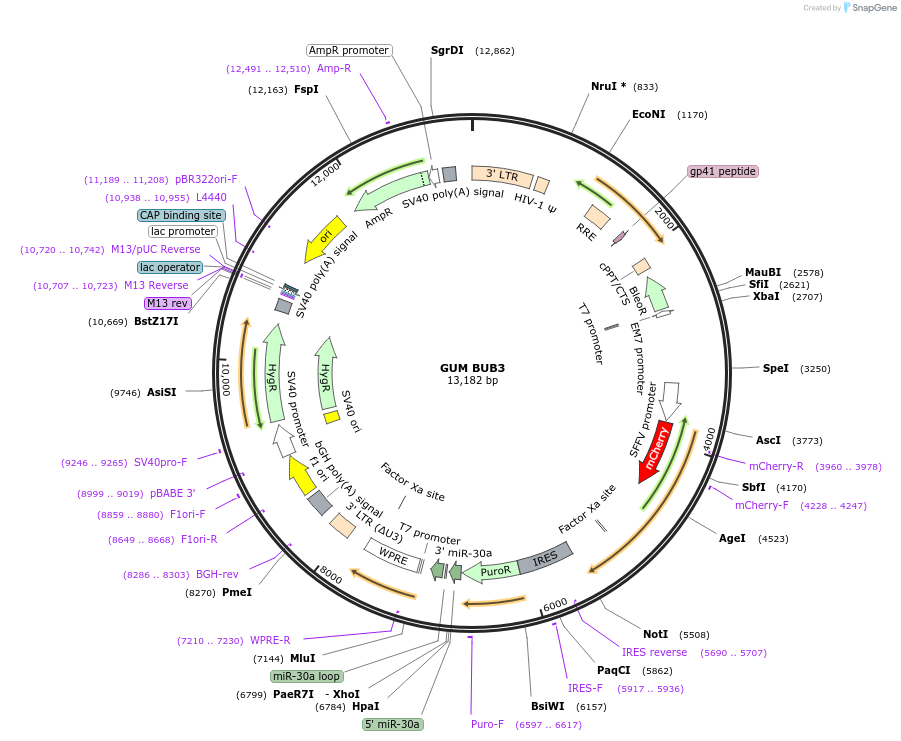
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 84029 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepGIPZ
-
Backbone manufacturerOpen Biosystems
- Backbone size w/o insert (bp) 10381
-
Vector typeMammalian Expression, Lentiviral
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin and Bleocin (Zeocin), 100 & 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameBUB3
-
Alt nameBUB3L
-
SpeciesH. sapiens (human)
-
Insert Size (bp)2801
-
GenBank ID9184
-
Entrez GeneBUB3 (a.k.a. BUB3L, hBUB3)
- Promoter UCOE / SFFV
-
Tag
/ Fusion Protein
- mCherry (N terminal on insert)
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer mCherry-F
- 3′ sequencing primer pCDH-rev
- (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byBUB3 ORF from Open Biosystems
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Parental vector (pGIPZ containing a non-silencing control hairpin) was cut with XbaI + NotI to remove the CMV promoter and TurboGFP. UCOE / SFFV enhancer promoter complex and ORF with N-terminal mCherry were PCR amplified and Gibson cloned into the cut vector.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
GUM BUB3 was a gift from Patrick Paddison (Addgene plasmid # 84029 ; http://n2t.net/addgene:84029 ; RRID:Addgene_84029) -
For your References section:
BuGZ is required for Bub3 stability, Bub1 kinetochore function, and chromosome alignment. Toledo CM, Herman JA, Olsen JB, Ding Y, Corrin P, Girard EJ, Olson JM, Emili A, DeLuca JG, Paddison PJ. Dev Cell. 2014 Feb 10;28(3):282-94. doi: 10.1016/j.devcel.2013.12.014. Epub 2014 Jan 23. 10.1016/j.devcel.2013.12.014 PubMed 24462187