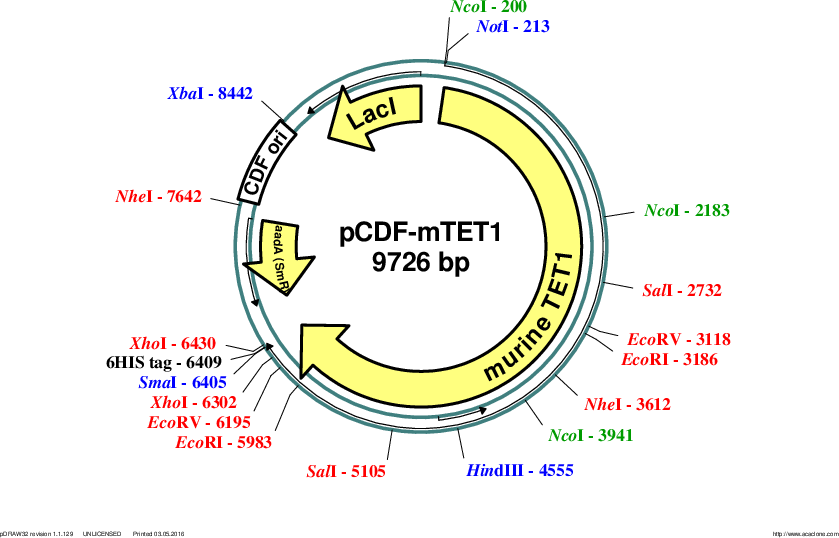
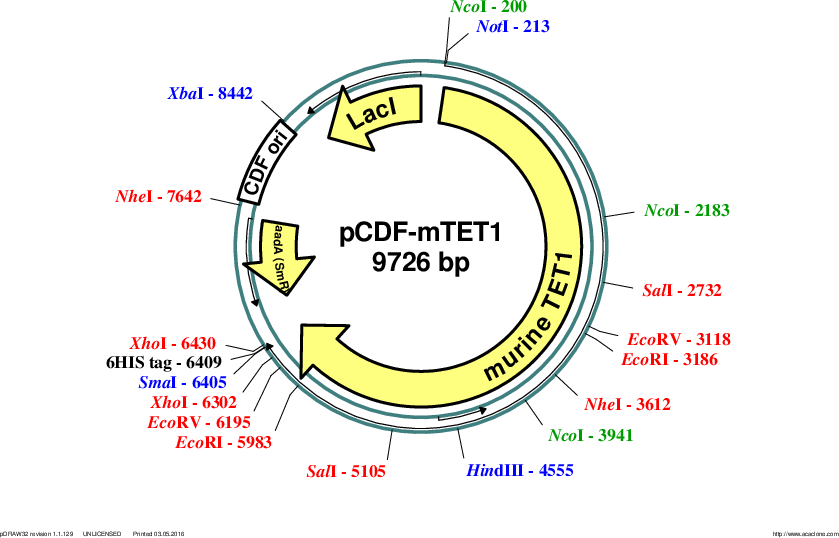
pCDF-mTET1
(Plasmid
#81052)
-
Purposebacterial expression of murine TET1
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 81052 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepCDF-Duet-1
-
Backbone manufacturerNovagen
- Backbone size w/o insert (bp) 3780
- Total vector size (bp) 9726
-
Modifications to backboneboth MCS were modified to generate NotI and SpeI cloning sites and to remove the N-terminal 6HIS-tag
-
Vector typeBacterial Expression
Growth in Bacteria
-
Bacterial Resistance(s)Streptomycin, 50 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)DH5alpha
-
Copy numberLow Copy
Gene/Insert
-
Gene/Insert nameTET1
-
SpeciesM. musculus (mouse)
-
Insert Size (bp)6171
-
GenBank IDNM_001253857.1
-
Entrez GeneTet1 (a.k.a. 2510010B09Rik, Cxxc6, D10Ertd17e, LCX, mKIAA1676)
- Promoter T7
-
Tag
/ Fusion Protein
- 6HIS (C terminal on insert)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site NotI (not destroyed)
- 3′ cloning site XbaI (destroyed during cloning)
- 5′ sequencing primer GGATCTCGACGCTCTCCCT
- 3′ sequencing primer T7_terminal_primer (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made bycDNA clone was a gift from Prof Radek Skoda, Department of Biomedicine, University of Basel
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCDF-mTET1 was a gift from Primo Schaer (Addgene plasmid # 81052 ; http://n2t.net/addgene:81052 ; RRID:Addgene_81052) -
For your References section:
Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Weber AR, Krawczyk C, Robertson AB, Kusnierczyk A, Vagbo CB, Schuermann D, Klungland A, Schar P. Nat Commun. 2016 Mar 2;7:10806. doi: 10.1038/ncomms10806. 10.1038/ncomms10806 PubMed 26932196