-
PurposeFusion protein Cas9+GFP
-
Depositing Labs
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 78546 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
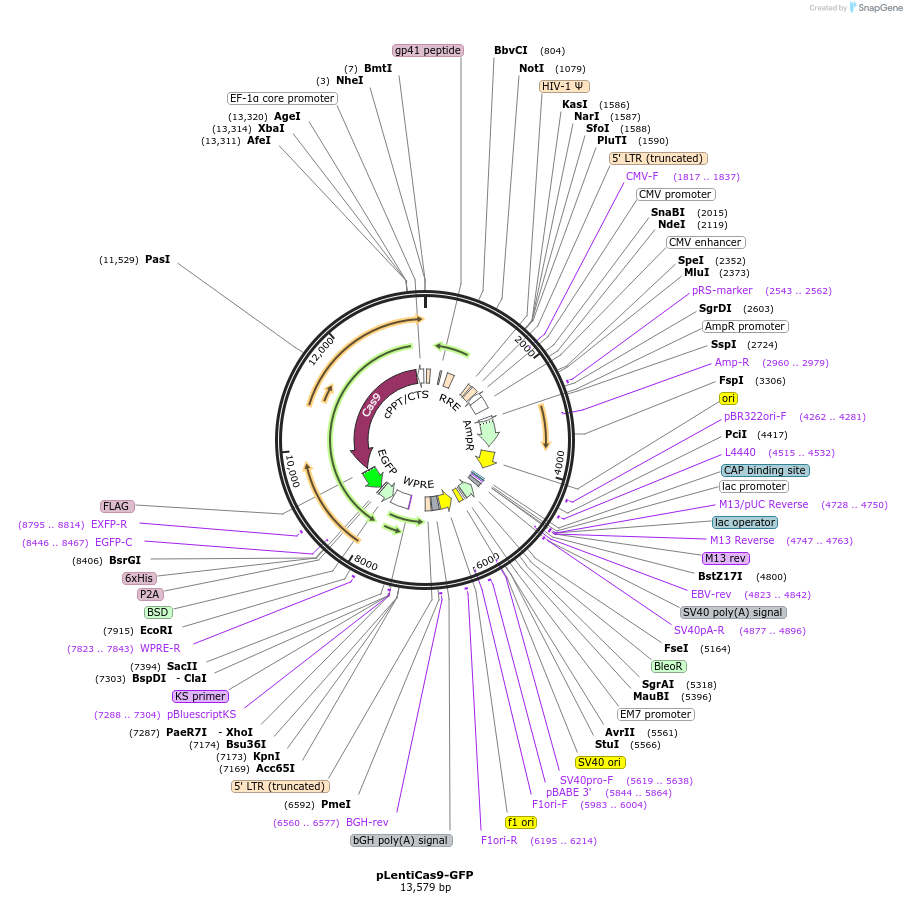
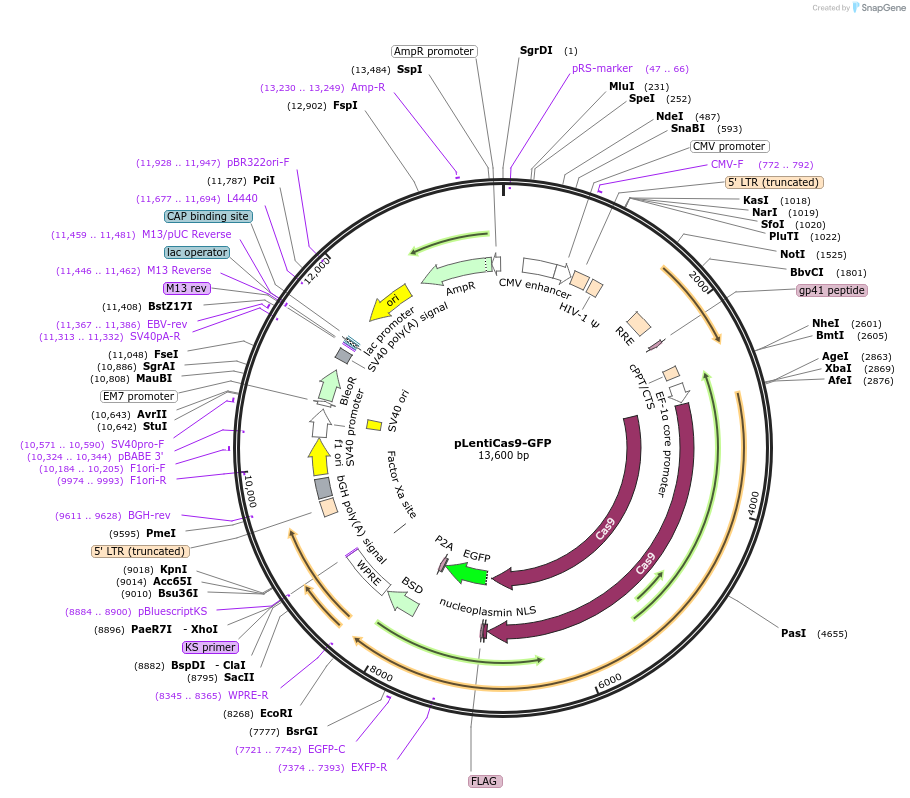
Vector backboneLenti-Cas9-blast
-
Backbone manufacturerAddgene 52962
- Backbone size w/o insert (bp) 12859
- Total vector size (bp) 13669
-
Modifications to backboneAdded GFP
-
Vector typeMammalian Expression, Lentiviral
-
Selectable markersBlasticidin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameGFP
-
Insert Size (bp)717
- Promoter Fusion protein with Cas9
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer CTGGACGCCACCCTGATCC
- 3′ sequencing primer Gacgctgtagtcttcagagatg
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Note: Compared to the provided full plasmid sequence, Addgene's quality control sequencing finds a G202S amino acid residue substitution in the EGFP fluorophore. The depositing laboratory is aware of this residue change, and it does not affect fluorophore function.
Note that, according to the depositing laboratory, this plasmid results in a weak GFP signal that may be difficult to see by microscope, but it is useful for flow cytometry.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pLentiCas9-GFP was a gift from Roderic Guigo & Rory Johnson (Addgene plasmid # 78546 ; http://n2t.net/addgene:78546 ; RRID:Addgene_78546) -
For your References section:
Scalable Design of Paired CRISPR Guide RNAs for Genomic Deletion. Pulido-Quetglas C, Aparicio-Prat E, Arnan C, Polidori T, Hermoso T, Palumbo E, Ponomarenko J, Guigo R, Johnson R. PLoS Comput Biol. 2017 Mar 2;13(3):e1005341. doi: 10.1371/journal.pcbi.1005341. eCollection 2017 Mar. PCOMPBIOL-D-16-01254 [pii] PubMed 28253259