-
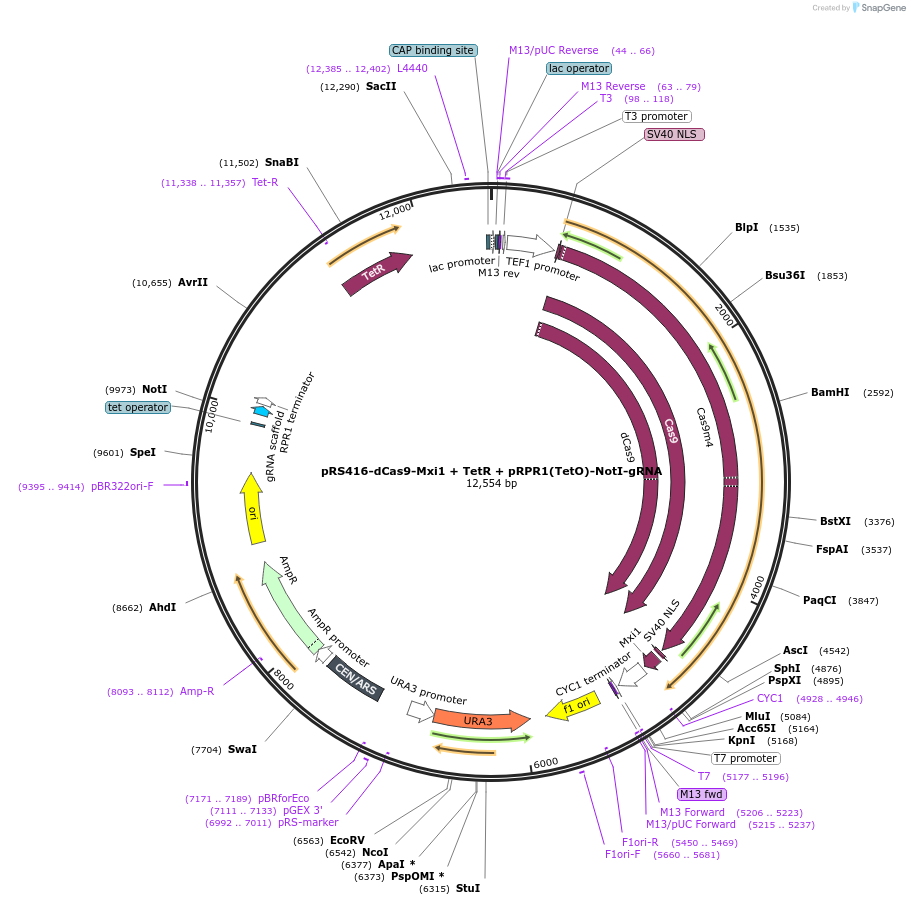
PurposepRS416 Ura marked Cen/Ars plasmid with dCas9-Mxi1 under Tef1 promoter, and tet-incucibile RPR1 promoter with NotI cloning site adjacent to gRNA
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 73796 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepRS416
-
Backbone manufacturerhttps://www.addgene.org/vector-database/3985/
- Backbone size w/o insert (bp) 4898
- Total vector size (bp) 12554
-
Modifications to backboneCut between multiple cloning site and PciI site outside of the multiple cloning site so part of vector is missing
-
Vector typeYeast Expression, CRISPR
-
Selectable markersURA3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberLow Copy
Gene/Insert 1
-
Gene/Insert namedCas9-Mxi1
-
SpeciesSynthetic; S pyogenes Cas9
-
Insert Size (bp)4344
-
MutationD10A, H840A
- Promoter pTef1
-
Tags
/ Fusion Proteins
- NLS (N terminal on insert)
- NLS (C terminal on insert)
- Mxi1 (C terminal on insert)
Cloning Information for Gene/Insert 1
- Cloning method Gibson Cloning
- 5′ sequencing primer CTCTTTCGATGACCTCCCATTG
- 3′ sequencing primer gtaatacgactcactataggg
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameTet Repressor
-
Alt nameTetR
-
SpeciesBacterial
-
Insert Size (bp)624
- Promoter pGPM1
Cloning Information for Gene/Insert 2
- Cloning method Gibson Cloning
- 5′ sequencing primer gagtacaaacgcatgaaatcc
- 3′ sequencing primer CGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAG
- (Common Sequencing Primers)
Gene/Insert 3
-
Gene/Insert nameStructural gRNA for S pyogenes
-
Alt namesgRNA
-
SpeciesSynthetic; S pyogenes
-
Insert Size (bp)82
- Promoter pRPR1(TetO)
Cloning Information for Gene/Insert 3
- Cloning method Gibson Cloning
- 5′ sequencing primer cagcaacgcggcctttttacggttcctggcc
- 3′ sequencing primer CAGGAAAGACCGCGGcttaaagtcatacattgcacgacta
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byCas9 gene was received from DiCarlo et al.
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Detailed benchling plasmid map available at: https://benchling.com/s/0WUKQB/edit
All annotations are available there. This is best reference to use for this plasmid. The genbank file included was exported from benchling for this plasmid.
Please note to clone in gRNAs you need to use Gibson Assembly. Gibson will chew back the NotI overhangs. Provide homology on either side of the cut site to clone. Generally we order 60mers with 20bp of homology to the promoter and to the terminator. For high efficiency amplify with primers that extend these homologies to 30-40bp. You can directly clone in single stranded oligo with 20bp overlaps using Gibson if you only need low efficiency. To do this use ~100x oligo to plasmid. For double stranded PCR product, use ratios between to 2:1 to 10:1 insert to plasmid. See paper methods for more details.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pRS416-dCas9-Mxi1 + TetR + pRPR1(TetO)-NotI-gRNA was a gift from Ronald Davis (Addgene plasmid # 73796 ; http://n2t.net/addgene:73796 ; RRID:Addgene_73796) -
For your References section:
Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Smith JD, Suresh S, Schlecht U, Wu M, Wagih O, Peltz G, Davis RW, Steinmetz LM, Parts L, St Onge RP. Genome Biol. 2016 Mar 8;17(1):45. doi: 10.1186/s13059-016-0900-9. 10.1186/s13059-016-0900-9 [pii] PubMed 26956608