-
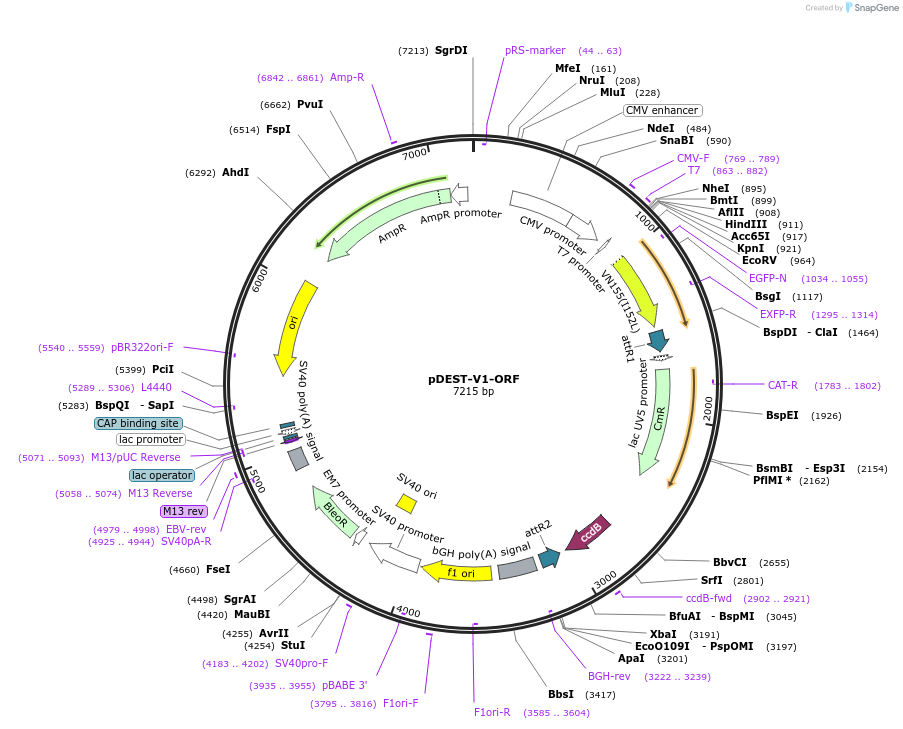
Purpose(Empty Backbone) Gateway™ destination vector encoding N-terminal fusion with Venus fluorescent protein fragment 1 (V1).
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 73635 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepcDNA3.1
-
Backbone manufacturerInvitrogen
- Backbone size (bp) 7215
-
Modifications to backboneInsertion of Gateway compatible recombination site. Insertion of Venus fluorescent protein fragment (V1) upstream and in frame of Gateway compatible recombination site.
-
Vector typeMammalian Expression
- Promoter CMV
-
Selectable markersZeocin
-
Tag
/ Fusion Protein
- Venus fragment 1 (V1) (N terminal on insert)
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol and Ampicillin, 25 & 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)ccdB Survival
-
Copy numberHigh Copy
Cloning Information
- Cloning method Restriction Enzyme
- 5′ sequencing primer CGCACCATCTTCTTCAAG
- 3′ sequencing primer TAGAAGGCACAGTCGAGG
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byVenus fragment was a kind gift from Prof. Stephen Michnick (University of Montreal). Plasmid built by Darren N. Saunders, Adrian Wan and Joseph Lau.
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pDEST-V1-ORF was a gift from Darren Saunders (Addgene plasmid # 73635 ; http://n2t.net/addgene:73635 ; RRID:Addgene_73635) -
For your References section:
Bimolecular complementation affinity purification (BiCAP) reveals dimer-specific protein interactions for ERBB2 dimers. Croucher DR, Iconomou M, Hastings JF, Kennedy SP, Han JZ, Shearer RF, McKenna J, Wan A, Lau J, Aparicio S, Saunders DN. Sci Signal. 2016 Jul 12;9(436):ra69. doi: 10.1126/scisignal.aaf0793. 10.1126/scisignal.aaf0793 PubMed 27405979