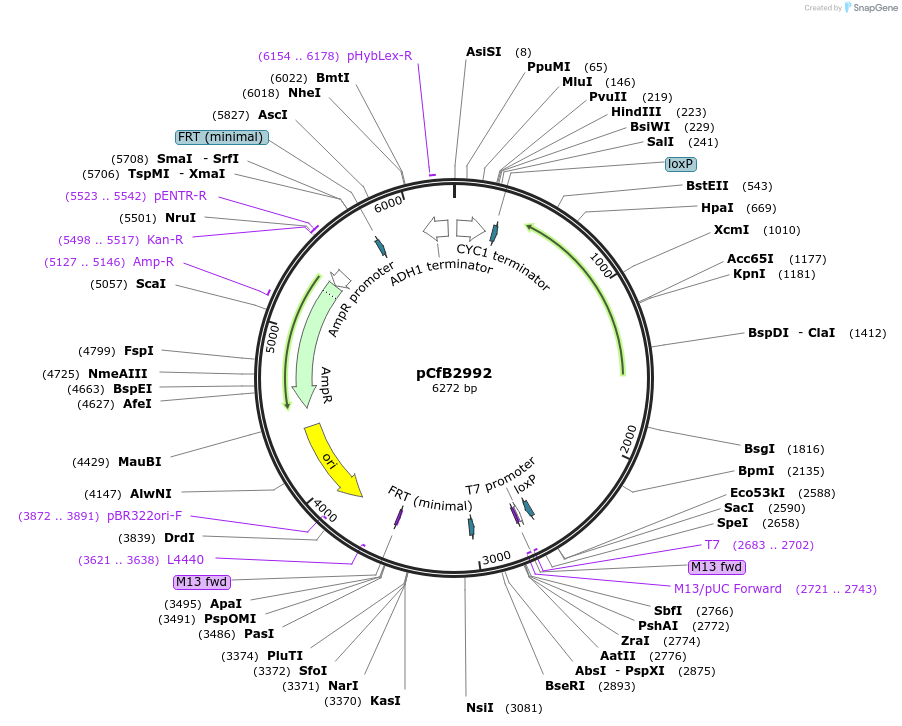
pCfB2992
(Plasmid
#63642)
-
Purpose(Empty Backbone) pCfB2992 is a vector for multiple integrations at sites sharing homology with Ty1Cons1. The selective marker is Kl.LEU2. Additionally it contains a USER cassette.
-
Depositing Labs
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 63642 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonesee publication
-
Vector typeYeast Expression, Cre/Lox
- Promoter none
-
Selectable markersLEU2
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Cloning Information
- Cloning method Unknown
- 5′ sequencing primer unknown
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
These vectors combine the advantage of efficient uracil excision reaction-based cloning (USER) and Cre-LoxP-mediated marker recycling system.
Please see previous FEMS Yeast Research paper for more details:
http://femsyr.oxfordjournals.org/content/14/2/238
Feature list:
USER cassette 1..14
T CYC1 20..190
loxP 257..316
T K.l. URA3 317..439
Degradation tag 440..490
K. l. LEU2 491..1576
P K. l. LEU2 1577..2606
loxP 2623..2656
Consensus TY1Cons1 3' 2796..3003
pUC ori 3776..4419
AmpR 4522..5380
Consensus TY1Cons1 5' 5824..6057
T ADH1 6084..6278
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pCfB2992 was a gift from Irina Borodina & Jerome Maury (Addgene plasmid # 63642 ; http://n2t.net/addgene:63642 ; RRID:Addgene_63642) -
For your References section:
EasyCloneMulti: A Set of Vectors for Simultaneous and Multiple Genomic Integrations in Saccharomyces cerevisiae. Maury J, Germann SM, Baallal Jacobsen SA, Jensen NB, Kildegaard KR, Herrgard MJ, Schneider K, Koza A, Forster J, Nielsen J, Borodina I. PLoS One. 2016 Mar 2;11(3):e0150394. doi: 10.1371/journal.pone.0150394. eCollection 2016. PONE-D-15-45506 [pii] PubMed 26934490