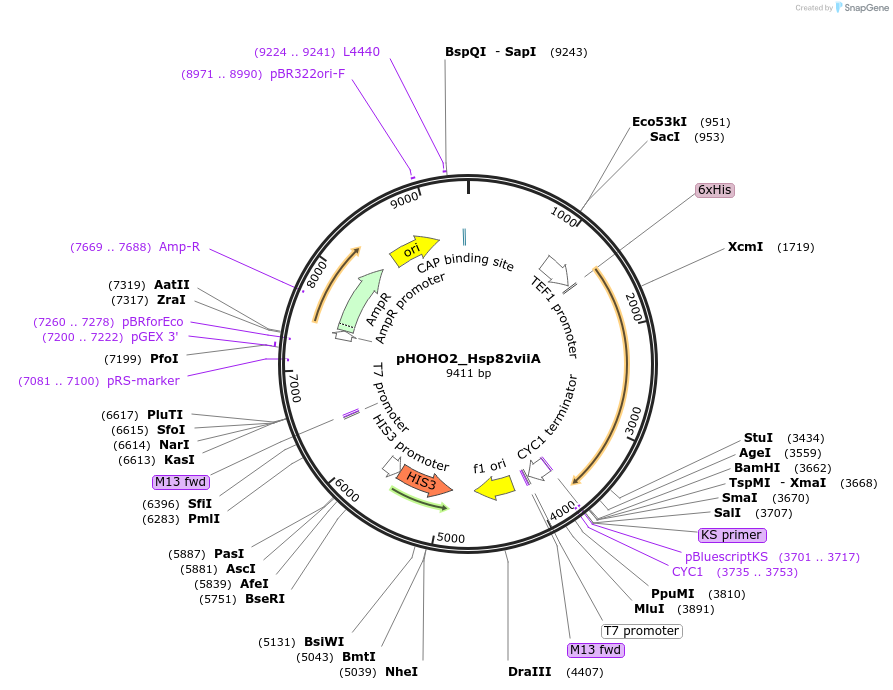
pHOHO2_Hsp82viiA
(Plasmid
#61708)
-
PurposeYeast integration plasmid targeted to the HO loci with heterodimeric Hsp82. Digest with NotI prior to addition to yeast.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 61708 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepHOHO2
-
Vector typeYeast Expression ; Yeast integration vector targeting inserts to the HO loci
-
Selectable markersHIS3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)XL1 Blue
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameHsp82viiA
-
Alt nameHsp90
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)2000
-
MutationHeteromeric coiled coil inserted after amino acid 678; A595C*
-
Entrez GeneHSP82 (a.k.a. YPL240C, HSP90)
- Promoter TEF1
-
Tag
/ Fusion Protein
- 6xHis (N terminal on insert)
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer CTCTTTCGATGACCTCCCATTG
- 3′ sequencing primer ctccttccttttcggttagag
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
*Note: This plasmid contains an A595C amino acid residue substitution. This residue change does not alter Hsp90 function in yeast but enables cross-dimer disulfide formation under oxidizing conditions. This construct accurately represents the plasmid as it was used in the associated publication.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pHOHO2_Hsp82viiA was a gift from Dan Bolon (Addgene plasmid # 61708 ; http://n2t.net/addgene:61708 ; RRID:Addgene_61708) -
For your References section:
Designed Hsp90 heterodimers reveal an asymmetric ATPase-driven mechanism in vivo. Mishra P, Bolon DN. Mol Cell. 2014 Jan 23;53(2):344-50. doi: 10.1016/j.molcel.2013.12.024. 10.1016/j.molcel.2013.12.024 PubMed 24462207