-
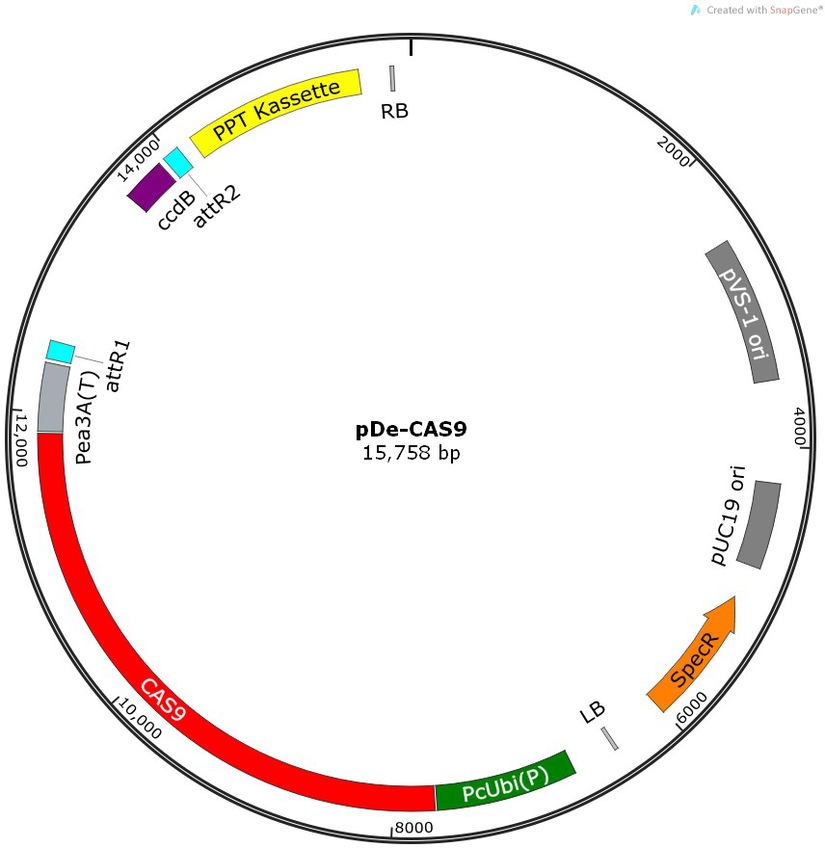
PurposeBinary expression vector with A.th. codon-optimized Cas9 and Gateway destination sequence
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 61433 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepPZP221
- Backbone size w/o insert (bp) 7100
- Total vector size (bp) 15800
-
Vector typePlant Expression, CRISPR
-
Selectable markersBasta
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol and Spectinomycin, 25 & 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DB3.1
-
Growth instructionscontains ccdB gene
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameCas9
-
SpeciesA. thaliana (mustard weed)
-
Insert Size (bp)4100
- Promoter PcUBI4-2
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site EcoRI (not destroyed)
- 3′ cloning site AvrII (not destroyed)
- 5′ sequencing primer M13rev
- 3′ sequencing primer M13fw (Common Sequencing Primers)
Resource Information
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pDe-CAS9 was a gift from Holger Puchta (Addgene plasmid # 61433 ; http://n2t.net/addgene:61433 ; RRID:Addgene_61433) -
For your References section:
Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Fauser F, Schiml S, Puchta H. Plant J. 2014 Jul;79(2):348-59. doi: 10.1111/tpj.12554. Epub 2014 Jun 17. 10.1111/tpj.12554 PubMed 24836556