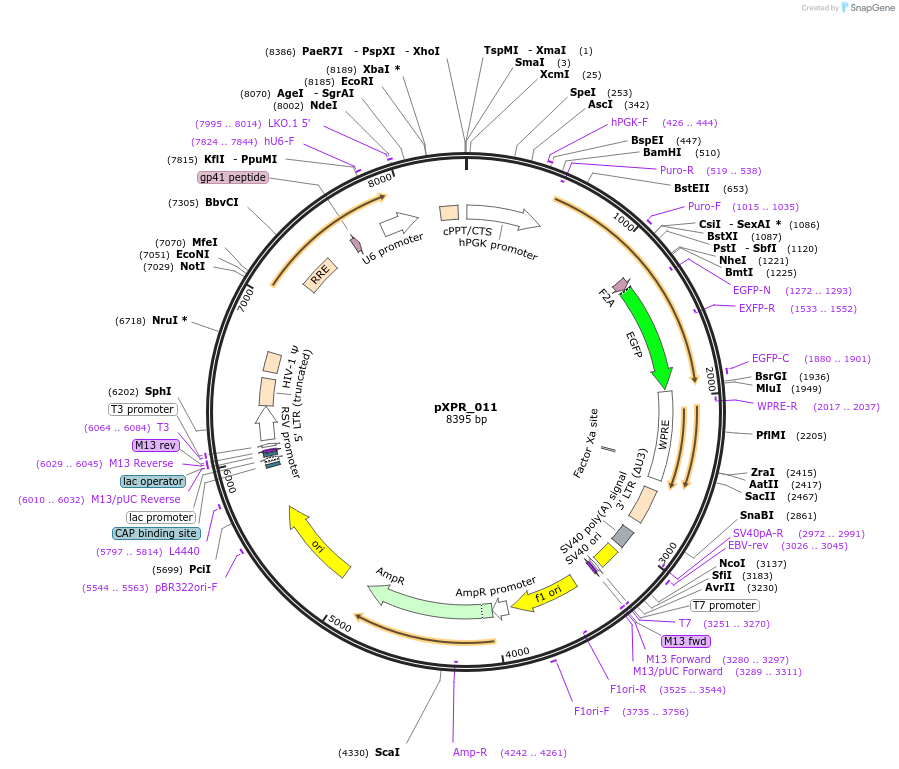
-
PurposeThis lentiviral vector can be used to assay Cas9 activity.
-
Depositing Labs
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 59702 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepLKO.1
- Total vector size (bp) 8395
-
Vector typeMammalian Expression, Lentiviral, CRISPR
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)Stbl3
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameEGFP sgRNA
-
SpeciesSynthetic
- Promoter hU6
Cloning Information
- Cloning method Unknown
- 5′ sequencing primer LKO.1 5' (Common Sequencing Primers)
Resource Information
-
Articles Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
This lentiviral vector can be used to assay Cas9 activity. In the absence of Cas9, the cells are both resistant to puromycin and express EGFP. In the presence of functional levels of Cas9, the sgRNA expressed from the U6 cassette targets EGFP, and thus cells will still be puromycin-resistant, but will no longer express EGFP. The fraction of EGFP-positive and -negative cells can be accurately quantitated by flow cytometry. Please note that it is unlikely that 100% of cells will ever be EGFP-negative, as a fraction of cells are likely to have in-frame indels, and those the cells may still be green. This assay is best done with cells infected with pXPR_011 at an MOI <1.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pXPR_011 was a gift from John Doench & David Root (Addgene plasmid # 59702 ; http://n2t.net/addgene:59702 ; RRID:Addgene_59702) -
For your References section:
Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Nat Biotechnol. 2014 Sep 3. doi: 10.1038/nbt.3026. 10.1038/nbt.3026 PubMed 25184501