-
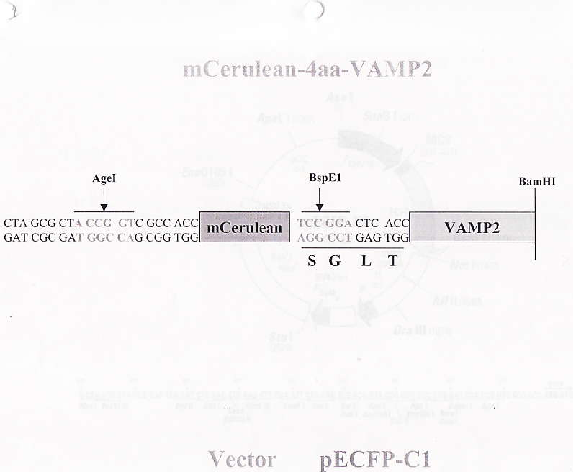
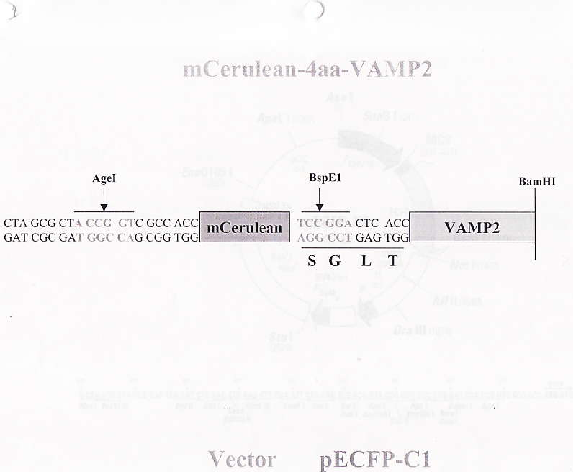
PurposeFluorescent probe: VAMP2 (synaptobrevin2), N-terminally-tagged with monomeric Cerulean, connected with a 4 amino acid linker. Expressed in mammalian cells, used with mCit-4aa-SNAP25B for FRET.
-
Depositing Lab
-
Sequence Information
Full plasmid sequence is not available for this item.
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 53233 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepECFP-C1
-
Backbone manufacturerClontech
- Backbone size w/o insert (bp) 4700
- Total vector size (bp) 5063
-
Modifications to backboneThis vector was created by mutagenesis of pECFP-C1. Compared to ECFP, Cerulean contains mutations: S72A, Y145A and H148D. mCerulean also contains A206K mutation to create a monomeric form of the fluorescent protein. See more details in Addgene's plasmid 27796.
-
Vector typeMammalian Expression
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameVAMP2
-
Alt nameVesicle-Associated Membrane Protein 2
-
Alt namesynaptobrevin 2
-
SpeciesR. norvegicus (rat)
-
Insert Size (bp)351
-
GenBank IDNM_012663
-
Entrez GeneVamp2 (a.k.a. RATVAMPB, RATVAMPIR, SYB, Syb2)
-
Tags
/ Fusion Proteins
- mCerulean (N terminal on backbone)
- 4aa-linker (N terminal on insert)
Resource Information
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
mCer-4aa-VAMP2 was a gift from Robert Zucker (Addgene plasmid # 53233 ; http://n2t.net/addgene:53233 ; RRID:Addgene_53233) -
For your References section:
Dance of the SNAREs: assembly and rearrangements detected with FRET at neuronal synapses. Degtyar V, Hafez IM, Bray C, Zucker RS. J Neurosci. 2013 Mar 27;33(13):5507-23. doi: 10.1523/JNEUROSCI.2337-12.2013. 10.1523/JNEUROSCI.2337-12.2013 PubMed 23536066
Map uploaded by the depositor.