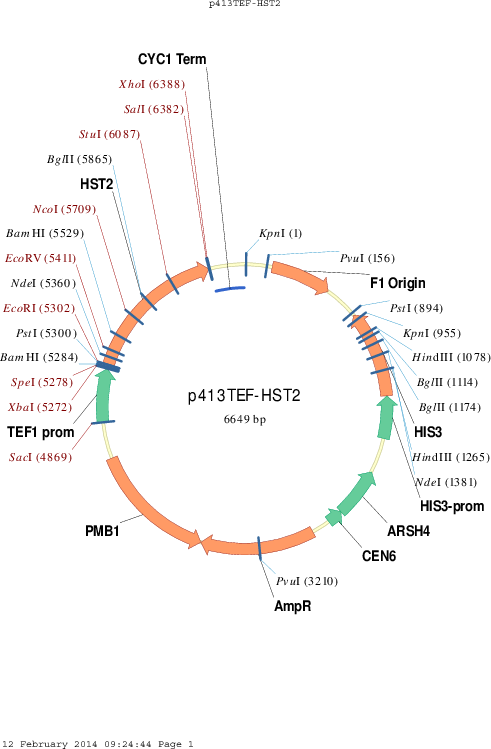
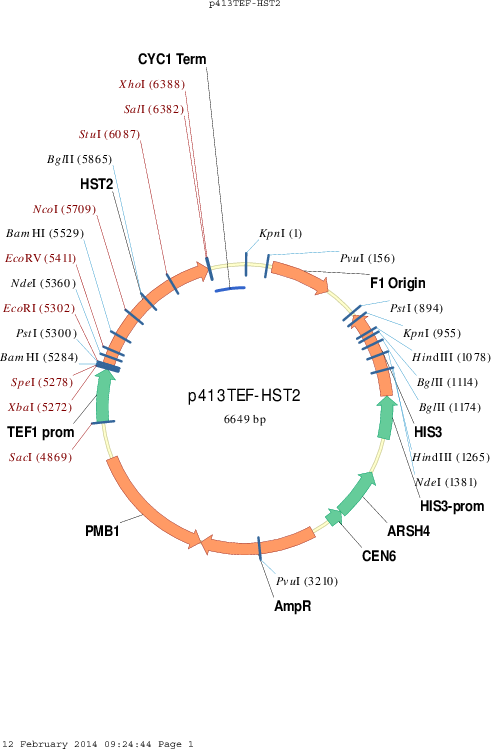
p413TEF-HST2
(Plasmid
#51745)
-
Purposeoverexpression of HST2 in S. cerevisiae, HIS3 marker
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 51745 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonep413TEF
-
Vector typeYeast Expression
-
Selectable markersHIS3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameHST2
-
Alt nameYPL015C
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)1074
-
Entrez GeneHST2 (a.k.a. YPL015C)
- Promoter TEF1
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site EcoRI (not destroyed)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer TEF1 (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
p413TEF-HST2 was a gift from Markus Ralser (Addgene plasmid # 51745 ; http://n2t.net/addgene:51745 ; RRID:Addgene_51745) -
For your References section:
Interfering with glycolysis causes Sir2-dependent hyper-recombination of Saccharomyces cerevisiae plasmids. Ralser M, Zeidler U, Lehrach H. PLoS One. 2009;4(4):e5376. doi: 10.1371/journal.pone.0005376. Epub 2009 Apr 24. 10.1371/journal.pone.0005376 PubMed 19390637