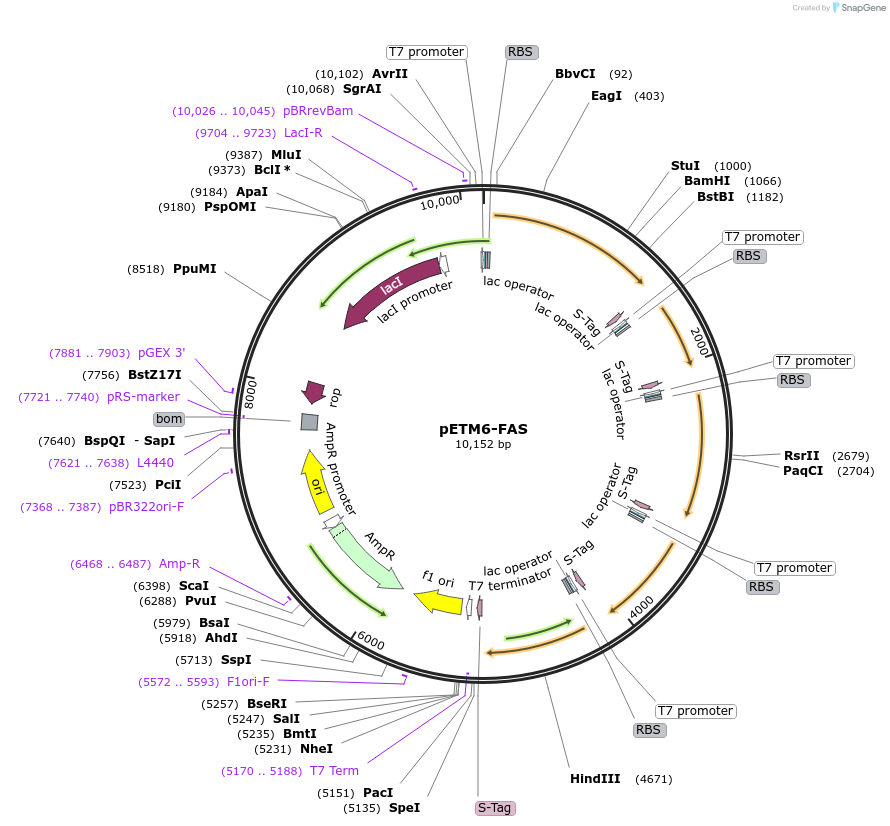
pETM6-FAS
(Plasmid
#49810)
-
PurposeEncodes CnfatB2, fabA, fabH, fabG, fabI in pseudo-operon configuration
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 49810 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepETM6
- Backbone size w/o insert (bp) 5203
- Total vector size (bp) 10152
-
Modifications to backboneSee paper.
-
Vector typeBacterial Expression
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert namefatty acyl-ACP thioesterase
-
Alt nameCnFatB2
-
SpeciesCocos nucifera
-
Insert Size (bp)1272
-
MutationCodon-optmized. See supplemental information of paper.
-
GenBank IDAEM72520.1
- Promoter T7
Cloning Information for Gene/Insert 1
- Cloning method Restriction Enzyme
- 5′ cloning site AvrII (destroyed during cloning)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer TGGTTGCGTCTATTGCC
- 3′ sequencing primer GTCGGACCCATATTGCCCAGA
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert namehydroxydecanoyl-ACP dehydratase
-
Alt namefabA
-
SpeciesE. coli K-12
-
Insert Size (bp)519
-
GenBank IDCDJ71394.1
- Promoter T7
Cloning Information for Gene/Insert 2
- Cloning method Restriction Enzyme
- 5′ cloning site AvrII (destroyed during cloning)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer GGTAGATAAACGCGAATCCTA
- 3′ sequencing primer CAGAAGGCAGACGTATCCTGG
- (Common Sequencing Primers)
Gene/Insert 3
-
Gene/Insert nameb-ketoacyl-ACP synthase III
-
Alt namefabH
-
SpeciesE. coli K-12
-
Insert Size (bp)954
-
GenBank IDEG10277
- Promoter T7
Cloning Information for Gene/Insert 3
- Cloning method Restriction Enzyme
- 5′ cloning site AvrII (destroyed during cloning)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer CACGAAGATTATTGGCACG
- 3′ sequencing primer AACGCACCAGAGCAGAAC
- (Common Sequencing Primers)
Gene/Insert 4
-
Gene/Insert name3-ketoacyl-ACP reductase
-
Alt namefabG
-
SpeciesE. coli K-12
-
Insert Size (bp)735
-
GenBank IDCDJ71528.1
- Promoter T7
Cloning Information for Gene/Insert 4
- Cloning method Restriction Enzyme
- 5′ cloning site AvrII (destroyed during cloning)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer ATCGCACTGGTAACCGG
- 3′ sequencing primer CAGACCATGTACATCCCGC
- (Common Sequencing Primers)
Gene/Insert 5
-
Gene/Insert nameenoyl-(acyl carrier protein) reductase
-
Alt namefabI
-
SpeciesE. coli K-12
-
Insert Size (bp)789
-
GenBank IDCDJ71704.1
- Promoter T7
Cloning Information for Gene/Insert 5
- Cloning method Restriction Enzyme
- 5′ cloning site AvrII (destroyed during cloning)
- 3′ cloning site SalI (not destroyed)
- 5′ sequencing primer GGGTTTTCTTTCCGGTAAGCGC
- 3′ sequencing primer CAGTTCGAGTTCGTTCATTGC
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pETM6-FAS was a gift from Mattheos Koffas (Addgene plasmid # 49810 ; http://n2t.net/addgene:49810 ; RRID:Addgene_49810) -
For your References section:
Modular optimization of multi-gene pathways for fatty acids production in E. coli. Xu P, Gu Q, Wang W, Wong L, Bower AG, Collins CH, Koffas MA. Nat Commun. 2013;4:1409. doi: 10.1038/ncomms2425. 10.1038/ncomms2425 PubMed 23361000