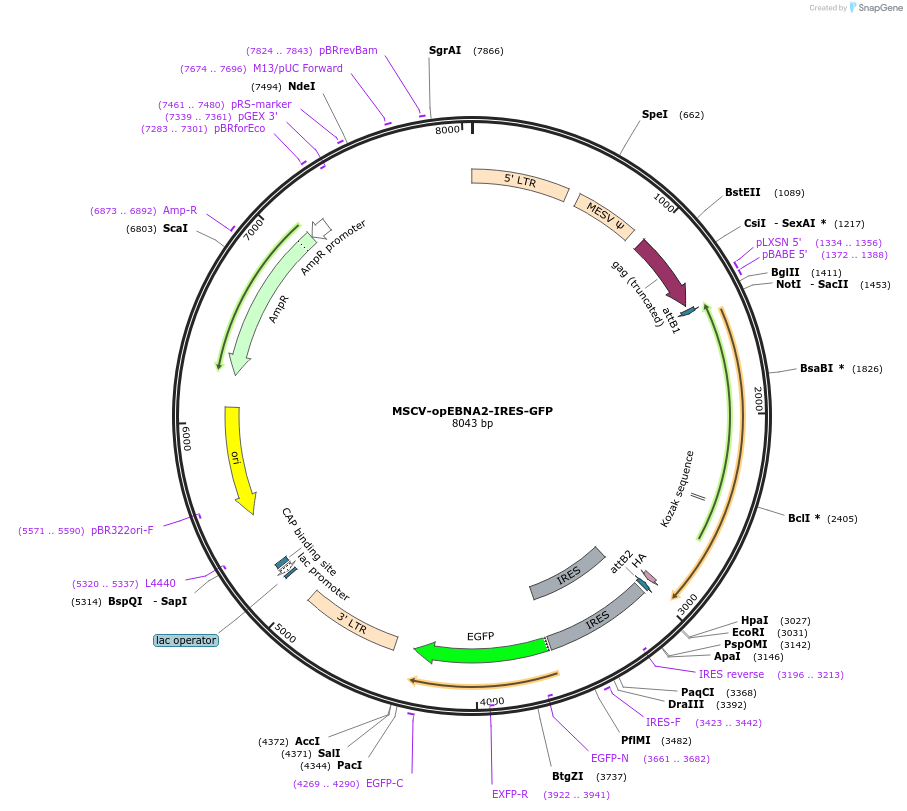
MSCV-opEBNA2-IRES-GFP
(Plasmid
#220354)
-
PurposeExpresses EBV latent gene EBNA2 in mammalian cells (EBNA2 coding sequence was optimised for expression and carries a C-terminal 3xHA-tag)
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 220354 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepMIGR1 (MSCV-IRES-GFP)
-
Vector typeMammalian Expression, Retroviral
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameopEBNA2
-
Alt nameexpression-optimised EBNA2 coding sequence
-
Alt nameexpression-optimised Epstein-Barr Nuclear Antigen 2 coding sequence
-
SpeciesEpstein-Barr Virus (EBV) strain B95-8
-
Insert Size (bp)1488
-
Mutationcodon-optimised for expression in mammalian cells
-
Entrez GeneEBNA-2 (a.k.a. HHV4_EBNA-2)
-
Tag
/ Fusion Protein
- 3 x HA tag (C terminal on insert)
Cloning Information
- Cloning method Gateway Cloning
- 5′ sequencing primer MSCV-gag Forward 5-cccttgaacctcctcgttcgacc-3
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
MSCV-opEBNA2-IRES-GFP was a gift from Klaus Rajewsky (Addgene plasmid # 220354 ; http://n2t.net/addgene:220354 ; RRID:Addgene_220354) -
For your References section:
LMP1 and EBNA2 constitute a minimal set of EBV genes for transformation of human B cells. Zhang J, Sommermann T, Li X, Gieselmann L, de la Rosa K, Stecklum M, Klein F, Kocks C, Rajewsky K. Front Immunol. 2023 Dec 19;14:1331730. doi: 10.3389/fimmu.2023.1331730. eCollection 2023. 10.3389/fimmu.2023.1331730 PubMed 38169736