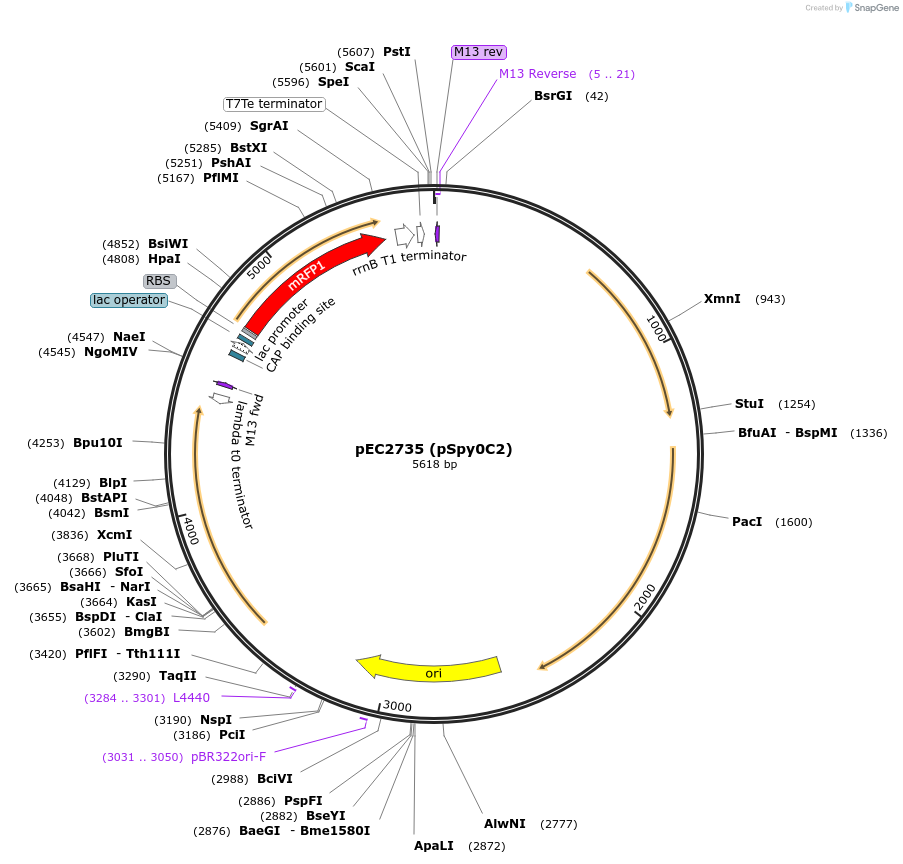
pEC2735 (pSpy0C2)
(Plasmid
#220279)
-
Purpose(Empty Backbone) integrative plasmid for heterologous gene expression in S. pyogenes SF370, integrates into the amyA locus via homologous recombination
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 220279 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepSpy0C2
- Backbone size (bp) 5618
-
Modifications to backboneSee associated publication for detailed construction information.
-
Vector typeBacterial Expression
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol, 25 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Growth instructionsFor E. coli, use 20 µg/mL chloramphenicol in LB medium. For S. pyogenes, also use 20 µg/mL chloramphenicol in THY medium. Successful integrations into the streptococcal genome can be detected on starch agar plates as integration into amyA will result in the inability to degrade starch.
-
Copy numberHigh Copy
Cloning Information
- Cloning method Golden Gate
- 5′ sequencing primer GTAAAACGACGGCCAGTC
- 3′ sequencing primer CAGGAAACAGCTATGAC
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
The multiple cloning site (EcoRI, XbaI, SpeI and PstI) contains the Plac_mrfp cassette. The mrfp reporter allows for red-white screening during sub-cloning in E. coli (DH5a). When the Plac_mrfp cassette is replaced with the insert of interest, colonies will appear white.
Resource Information: The MCS featuring the mrfp cassette and the chloramphenicol resistance cassette were taken from pSpy1C. The pUC origin for replication in E. coli was taken from pUC19-mKate2∼ssrA. Up- and downstream homologous regions of amyA were amplified from the S. pyogenes SF370 genome.
Linearise the plasmid with the restriction enzyme Alw44I before transformation into S. pyogenes to favour double crossover recombination of the plasmid into the amyA locus. Positive transformants will show loss in starch degradation activity on starch agar plates.
Please download the file linked below to obtain a well annotated map of the plasmid.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pEC2735 (pSpy0C2) was a gift from Emmanuelle Charpentier (Addgene plasmid # 220279 ; http://n2t.net/addgene:220279 ; RRID:Addgene_220279) -
For your References section:
Expanding the genetic toolbox for the obligate human pathogen Streptococcus pyogenes. Lautenschlager N, Schmidt K, Schiffer C, Wulff TF, Hahnke K, Finstermeier K, Mansour M, Elsholz AKW, Charpentier E. Front Bioeng Biotechnol. 2024 Jun 7;12:1395659. doi: 10.3389/fbioe.2024.1395659. eCollection 2024. 10.3389/fbioe.2024.1395659 PubMed 38911550