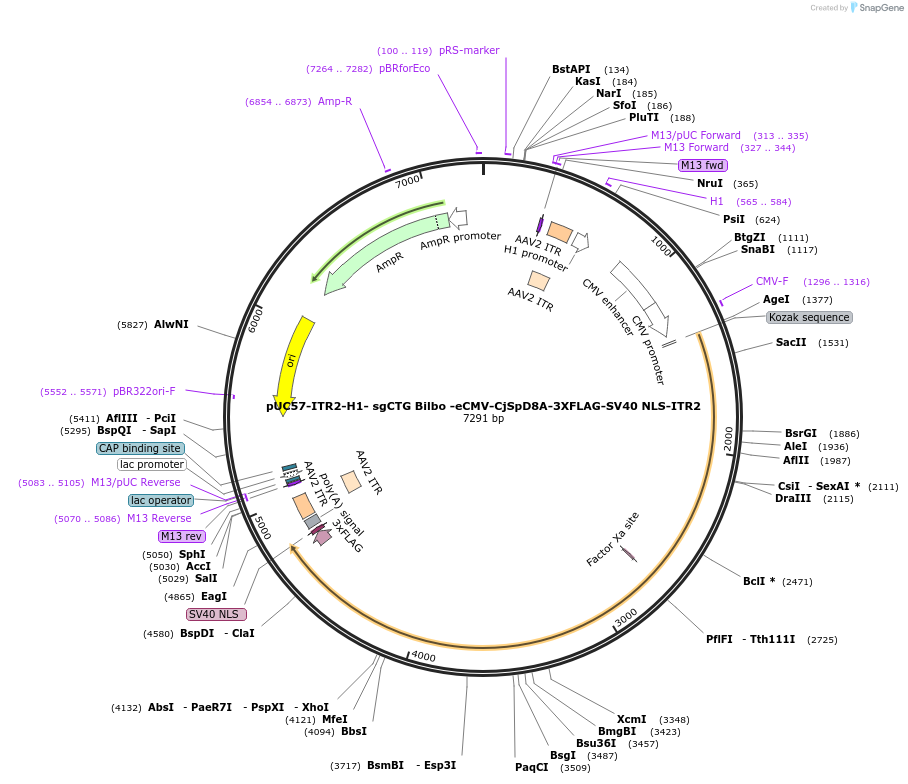
pUC57-ITR2-H1- sgCTG Bilbo -eCMV-CjSpD8A-3XFLAG-SV40 NLS-ITR2
(Plasmid
#210749)
-
PurposeCoding for CjSp D8A alongside Bilbo sgRNA targeting CAG repeats
-
Depositing Lab
-
Publication
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 210749 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepUC57
-
Backbone manufacturerGenscript
-
Vector typeMammalian Expression, CRISPR
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameCjSp D8A Cas9
-
SpeciesSynthetic
-
MutationD8A, N-terminal Cj Cas9, C-terminal Sp Cas9
- Promoter eCMV
-
Tag
/ Fusion Protein
- 3xFLAG, SV40 NLS, PolyA signal (C terminal on insert)
Cloning Information for Gene/Insert 1
- Cloning method Unknown
- 5′ sequencing primer n/a
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameBilbo sgCAG
-
Alt namegRNA target: GCTGCTGCTGCTGCTGCTGGTTTTAGTCCCTGAAAAGGGACTAAAATGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCT
- Promoter H1
Cloning Information for Gene/Insert 2
- Cloning method Unknown
- 5′ sequencing primer n/a
- (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byGenscript
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please visit https://doi.org/10.1101/2023.10.24.563270 for bioRxiv preprint.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pUC57-ITR2-H1- sgCTG Bilbo -eCMV-CjSpD8A-3XFLAG-SV40 NLS-ITR2 was a gift from Vincent Dion (Addgene plasmid # 210749 ; http://n2t.net/addgene:210749 ; RRID:Addgene_210749) -
For your References section:
Developing small Cas9 hybrids using molecular modeling. Mangin A, Dion V, Menzies G. bioRxiv 2023.10.24.563270 10.1101/2023.10.24.563270