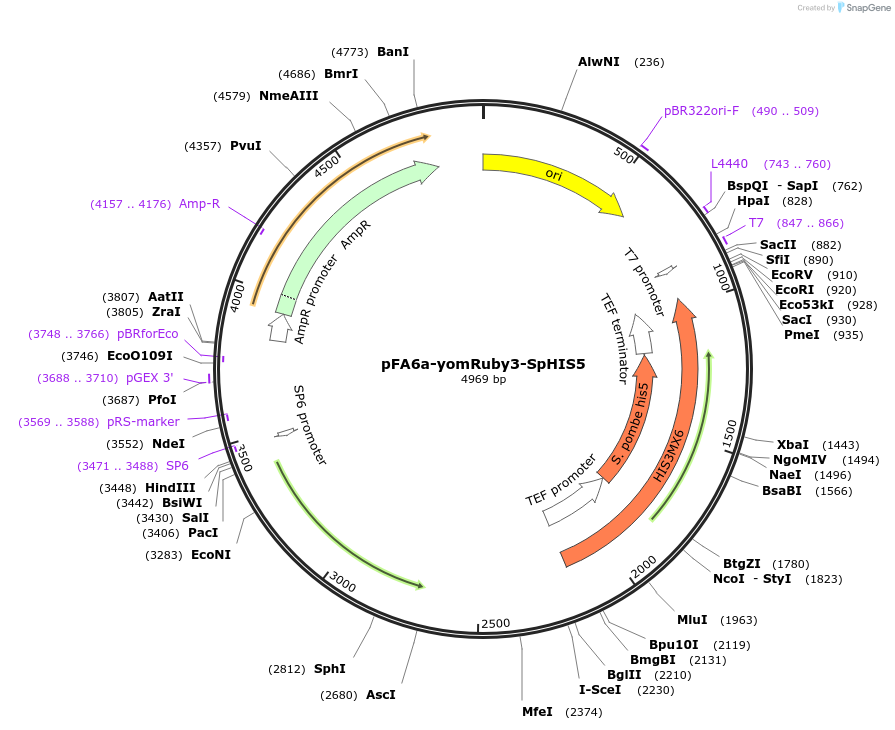
pFA6a-yomRuby3-SpHIS5
(Plasmid
#204287)
-
PurposeFor C-terminal tagging using yeast optimized mRuby3
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 204287 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepFA-Envy-SpHIS5 (Addgene plasmid 60782)
- Backbone size w/o insert (bp) 4021
- Total vector size (bp) 4969
-
Modifications to backboneWe replaced the Envy fluorescent marker with yomRuby3, which we had synthesized. We aslo added the meiotic high capacity terminator from the DIT1 gene at the 3' end
-
Vector typeYeast Expression
-
Selectable markersS pombe his5
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert namepFA6-yomRuby3-SpHIS5
-
Alt nameyomRuby3 for C-terminal tagging
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)1057
- Promoter none
-
Tag
/ Fusion Protein
- yomRuby3 (C terminal on backbone)
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer SP6
- 3′ sequencing primer GGG TAT TCT GGG CCT CCA TGT C (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byWe synthesized yomRuby3 (Twist Bioscience)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pFA6a-yomRuby3-SpHIS5 was a gift from Linda Huang (Addgene plasmid # 204287 ; http://n2t.net/addgene:204287 ; RRID:Addgene_204287) -
For your References section:
Meiosis II spindle disassembly requires two distinct pathways. Seitz BC, Mucelli X, Majano M, Wallis Z, Dodge AC, Carmona C, Durant M, Maynard S, Huang LS. Mol Biol Cell. 2023 Sep 1;34(10):ar98. doi: 10.1091/mbc.E23-03-0096. Epub 2023 Jul 12. 10.1091/mbc.E23-03-0096 PubMed 37436806