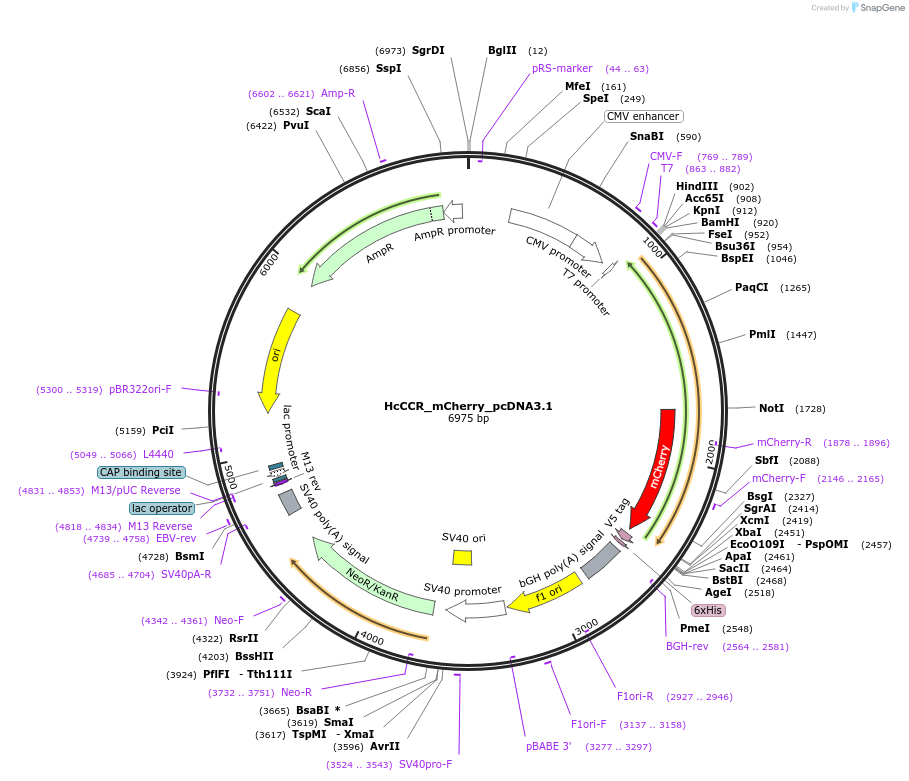
HcCCR_mCherry_pcDNA3.1
(Plasmid
#201687)
-
Purposeexpresses HcCCR_mCherry fusion in mammalian cells
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 201687 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepcDNA3.1
-
Backbone manufacturerInvitrogen
- Backbone size w/o insert (bp) 5428
- Total vector size (bp) 6940
-
Vector typeMammalian Expression
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameHcCCR
-
SpeciesHyphochytrium catenoides
-
Insert Size (bp)795
-
GenBank IDUTD53596 UTD53596
-
Tag
/ Fusion Protein
- mCherry (C terminal on backbone)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BamHI (not destroyed)
- 3′ cloning site NotI (not destroyed)
- 5′ sequencing primer T7 (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
HcCCR_mCherry_pcDNA3.1 was a gift from John Spudich (Addgene plasmid # 201687 ; http://n2t.net/addgene:201687 ; RRID:Addgene_201687) -
For your References section:
Structural Foundations of Potassium Selectivity in Channelrhodopsins. Govorunova EG, Sineshchekov OA, Brown LS, Bondar AN, Spudich JL. mBio. 2022 Dec 20;13(6):e0303922. doi: 10.1128/mbio.03039-22. Epub 2022 Nov 22. 10.1128/mbio.03039-22 PubMed 36413022