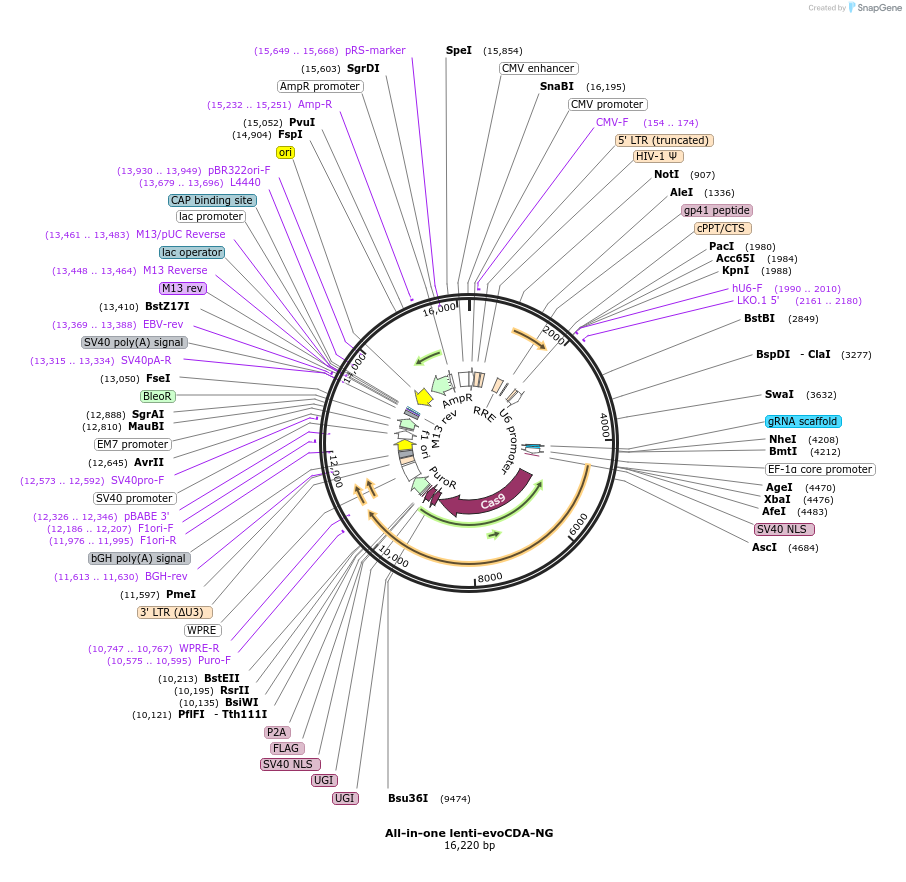
All-in-one lenti-evoCDA-NG
(Plasmid
#200449)
-
Purpose(Empty Backbone) Lentivirus vector for base editing
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 200449 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonelentiCRISPR v2
-
Modifications to backboneInserted base editor (#125613) to lentivirus plasmid #52961.
-
Vector typeMammalian Expression, Lentiviral, CRISPR
-
Selectable markersPuromycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)NEB Stable
-
Copy numberLow Copy
Cloning Information
- Cloning method Ligation Independent Cloning
- 5′ sequencing primer GACTATCATATGCTTACCGT
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byBase editor (#125613, gift from David Liu, Broad Institute) was inserted into lentivirus plasmid (#52961, gift from Feng Zhang, Broad Institute).
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please visit https://www.biorxiv.org/content/10.1101/2022.11.17.516964v1 for bioRxiv preprint.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
All-in-one lenti-evoCDA-NG was a gift from Benjamin Cravatt (Addgene plasmid # 200449 ; http://n2t.net/addgene:200449 ; RRID:Addgene_200449) -
For your References section:
Assigning functionality to cysteines by base editing of cancer dependency genes. Li H, Ma T, Remsberg JR, Won SJ, DeMeester KE, Njomen E, Ogasawara D, Zhao KT, Huang TP, Lu B, Simon GM, Melillo B, Schreiber SL, Lykke-Andersen J, Liu DR, Cravatt BF. Nat Chem Biol. 2023 Nov;19(11):1320-1330. doi: 10.1038/s41589-023-01428-w. Epub 2023 Oct 2. 10.1038/s41589-023-01428-w PubMed 37783940