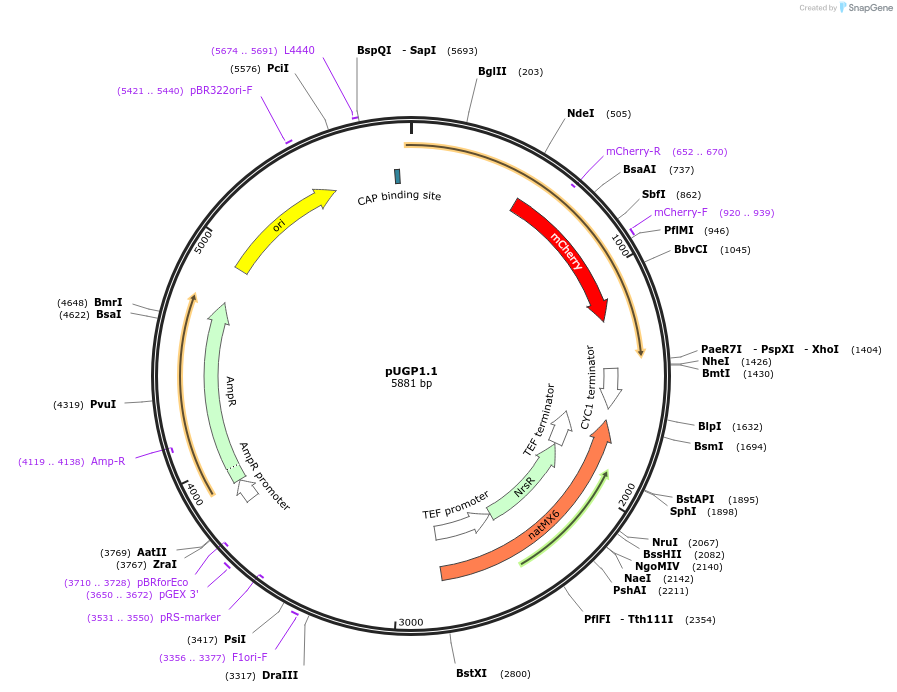
pUGP1.1
(Plasmid
#196615)
-
PurposePlasmid used for the C-terminal tagging of Ugp1 with mCherry and the auxin-inducible degron in yeast
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 196615 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepG23A, originally HO-poly-KanMX4-HO
-
Backbone manufacturerhttps://www.ncbi.nlm.nih.gov/pmc/articles/PMC55758/
- Backbone size w/o insert (bp) 2531
- Total vector size (bp) 5881
-
Vector typeYeast Expression
-
Selectable markersNatMX (select with Nourseotrichin)
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameUGP1::mCherry-AID(71-114)-NatMX
-
Alt nameYKL035W
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)3350
-
Entrez GeneUGP1 (a.k.a. YKL035W)
- Promoter Native promoter
-
Tags
/ Fusion Proteins
- mCherry (C terminal on insert)
- AID(71-114) (C terminal on insert)
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer CTGTCAAGGAGGGTATTCTGG
- 3′ sequencing primer CCTTGAAGCGCATGAACTC
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Cloning details are provided in Supplementary Table 2 of the paper. Details of the parent plasmid pG23A can be found on Addgene (Plasmid #102884).
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pUGP1.1 was a gift from Matthias Heinemann (Addgene plasmid # 196615 ; http://n2t.net/addgene:196615 ; RRID:Addgene_196615) -
For your References section:
Temporal segregation of biosynthetic processes is responsible for metabolic oscillations during the budding yeast cell cycle. Takhaveev V, Ozsezen S, Smith EN, Zylstra A, Chaillet ML, Chen H, Papagiannakis A, Milias-Argeitis A, Heinemann M. Nat Metab. 2023 Feb;5(2):294-313. doi: 10.1038/s42255-023-00741-x. Epub 2023 Feb 27. 10.1038/s42255-023-00741-x PubMed 36849832