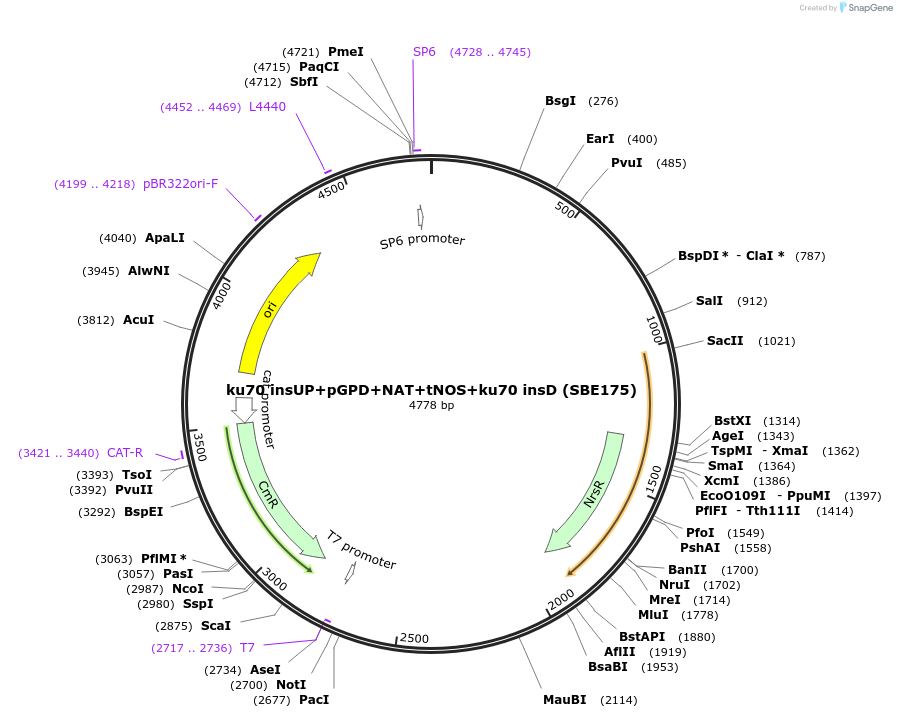
ku70 insUP+pGPD+NAT+tNOS+ku70 insD (SBE175)
(Plasmid
#195051)
-
Purposeintegration cassette for ku70 deletion and expression of NAT gene under pGPD for characterization
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 195051 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepGGA
- Backbone size w/o insert (bp) 2137
- Total vector size (bp) 4778
-
Vector typeBacterial Expression
-
Selectable markersNAT
Growth in Bacteria
-
Bacterial Resistance(s)Chloramphenicol, 25 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameNAT
-
SpeciesRhodotorula toruloides
-
Insert Size (bp)2655
- Promoter pGPD
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BsaI (destroyed during cloning)
- 3′ cloning site BsaI (destroyed during cloning)
- 5′ sequencing primer atttaggtgacactatag
- 3′ sequencing primer taatacgactcactataggg
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
ku70 insUP+pGPD+NAT+tNOS+ku70 insD (SBE175) was a gift from Petri-Jaan Lahtvee (Addgene plasmid # 195051 ; http://n2t.net/addgene:195051 ; RRID:Addgene_195051) -
For your References section:
Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides. Bonturi N, Pinheiro MJ, de Oliveira PM, Rusadze E, Eichinger T, Liudziute G, De Biaggi JS, Brauer A, Remm M, Miranda EA, Ledesma-Amaro R, Lahtvee PJ. Metab Eng Commun. 2022 May 23;15:e00200. doi: 10.1016/j.mec.2022.e00200. eCollection 2022 Dec. 10.1016/j.mec.2022.e00200 PubMed 35662893