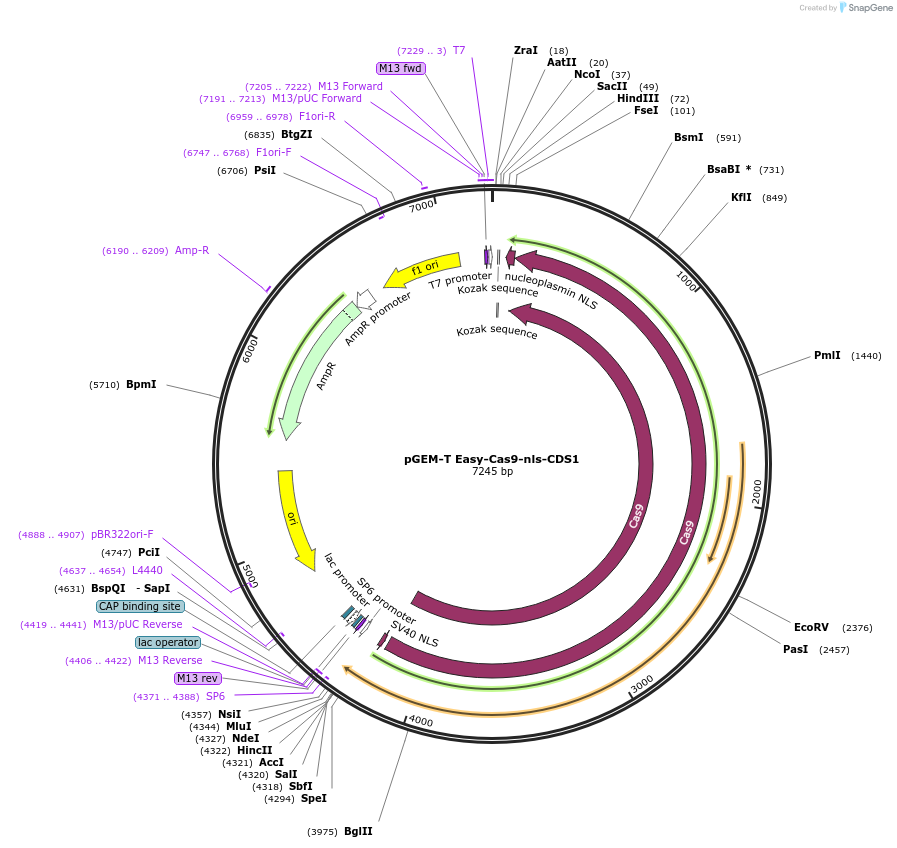
pGEM-T Easy-Cas9-nls-CDS1
(Plasmid
#160231)
-
PurposeCas9-nuclear localization, level 0 of MoClo Golden Gate position CDS1
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 160231 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepGEMT -Easy
- Backbone size w/o insert (bp) 3015
- Total vector size (bp) 7245
-
Vector typelevel 0 of MoClo Golden Gate position CDS1
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameCas9-nuclear localization signal gene sequence, Golden Gate MoClo position CDS1
-
Insert Size (bp)4199
Cloning Information
- Cloning method TOPO Cloning
- 5′ sequencing primer AATTCGATTATTGGTCTCTAAG
- 3′ sequencing primer GGCCGCGAATTCACTAGTGAT
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pGEM-T Easy-Cas9-nls-CDS1 was a gift from Matias Kirst (Addgene plasmid # 160231 ; http://n2t.net/addgene:160231 ; RRID:Addgene_160231) -
For your References section:
Simple, efficient and open-source CRISPR/Cas9 strategy for multi-site genome editing in Populus tremula x alba. Triozzi P, Schmidt HW, Dervinis C, Kirst M, Conde D. Tree Physiol. 2021 May 7. pii: 6270869. doi: 10.1093/treephys/tpab066. 10.1093/treephys/tpab066 PubMed 33960379