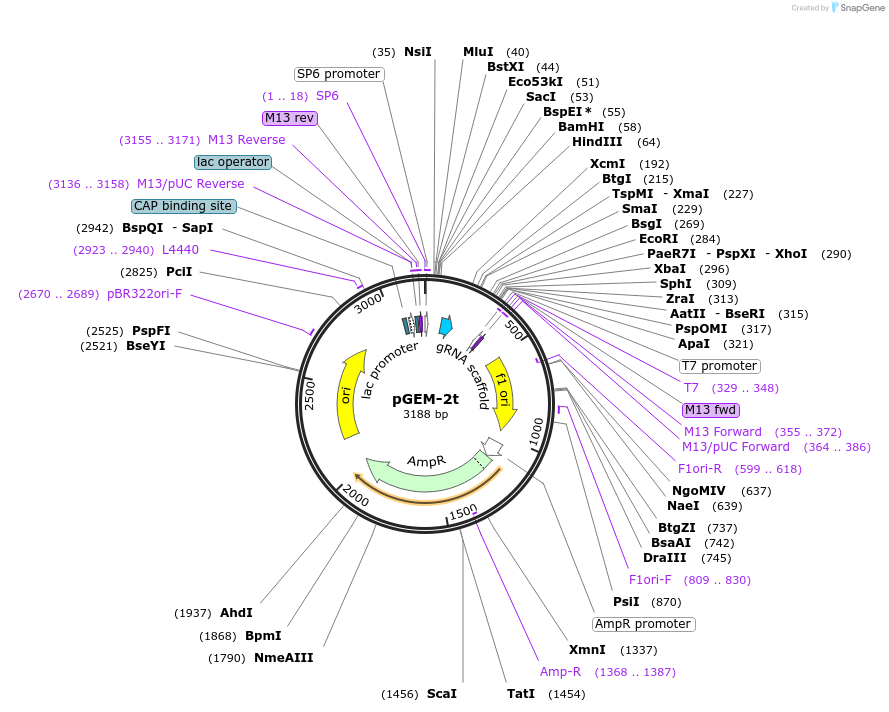
pGEM-2t
(Plasmid
#159752)
-
PurposeCRISPR/Cas9 genome editing in plants; a template for PCR-derived fragments used in the assembly of polycistronic transcripts, where sgRNAs are separated by an Arabidopsis alanine tRNA.
-
Depositing Lab
-
Publication
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 159752 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 * | |
* Log in to view industry pricing.
Backbone
-
Vector backbonepGEM7Zf
-
Backbone manufacturerPromega
-
Vector typeCRISPR
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert namesgRNA
-
gRNA/shRNA sequenceGTTTCTCACGCGAGCAATTC
-
SpeciesA. thaliana (mustard weed)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site HindIII (not destroyed)
- 3′ cloning site EcoRI (not destroyed)
- 5′ sequencing primer SP6
- 3′ sequencing primer T7
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please visit https://www.biorxiv.org/content/10.1101/2020.08.16.253203v1 for BioRxiv preprint
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pGEM-2t was a gift from Wolfgang Lukowitz (Addgene plasmid # 159752 ; http://n2t.net/addgene:159752 ; RRID:Addgene_159752) -
For your References section:
Targeted mutagenesis of the Arabidopsis GROWTH-REGULATING FACTOR (GRF) gene family suggests competition of multiplexed sgRNAs for Cas9 apoprotein. Angulo J, Astin CP, Bauer O , Blash KJ, Bowen NM, Chukwudinma NJ, Dinofrio AS, Faletti DO, Ghulam AM, Gusinde-Duffy CM, Horace KJ, Ingram AM, Isaack KE, Kiser RJ, Kobylanski JS, Long MR, Manning GA, Morales JM, Nguyen KH, Pham RT, Phillips MH, Reel TW, Seo JE, Vo HD, Wukuson AM, Yeary KA, Zheng GY, Lukowitz W. bioRxiv 10.1101/2020.08.16.253203