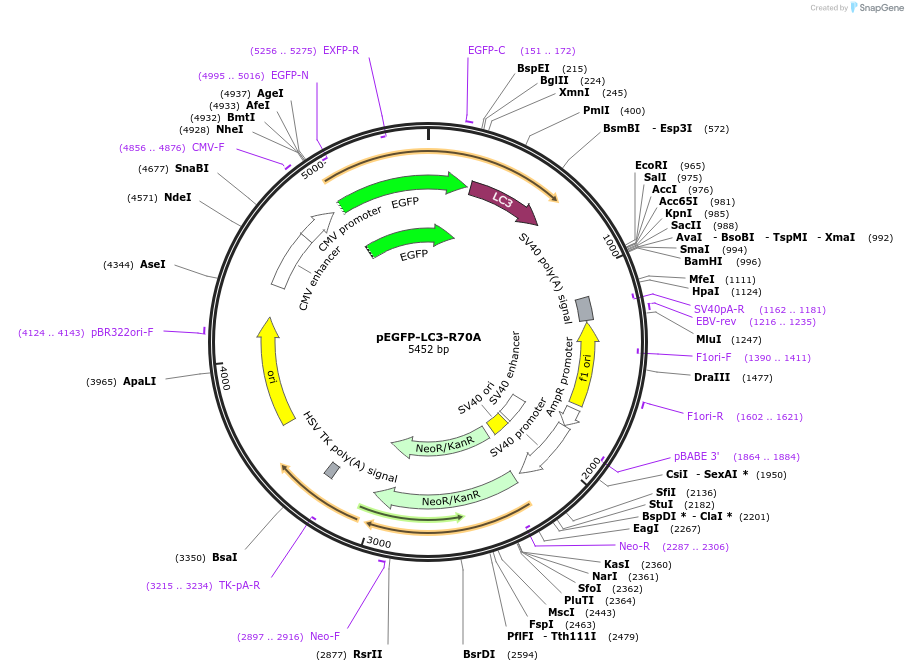
pEGFP-LC3-R70A
(Plasmid
#155264)
-
PurposeMammalian expression of rat LC3 fused to EGFP
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 155264 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepEGFP-C1
-
Backbone manufacturerclontech
-
Vector typeMammalian Expression
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert namemicrotubule-associated protein 1 light chain 3 beta
-
SpeciesR. norvegicus (rat)
-
MutationArginine 70 to Alanine
-
Entrez GeneMap1lc3b (a.k.a. LC3B, Map1lc3, Mpl3, zbs559)
-
Tag
/ Fusion Protein
- pEGFP (N terminal on backbone)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BglII (unknown if destroyed)
- 3′ cloning site EcoRI (unknown if destroyed)
- 5′ sequencing primer unknown (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made bymicrotubule-associated protein 1 light chain 3 beta (mutated from Addgene Plasmid #21073)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pEGFP-LC3-R70A was a gift from David Rubinsztein (Addgene plasmid # 155264 ; http://n2t.net/addgene:155264 ; RRID:Addgene_155264) -
For your References section:
A DNM2 Centronuclear Myopathy Mutation Reveals a Link between Recycling Endosome Scission and Autophagy. Puri C, Manni MM, Vicinanza M, Hilcenko C, Zhu Y, Runwal G, Stamatakou E, Menzies FM, Mamchaoui K, Bitoun M, Rubinsztein DC. Dev Cell. 2020 Apr 20;53(2):154-168.e6. doi: 10.1016/j.devcel.2020.03.018. 10.1016/j.devcel.2020.03.018 PubMed 32315611