-
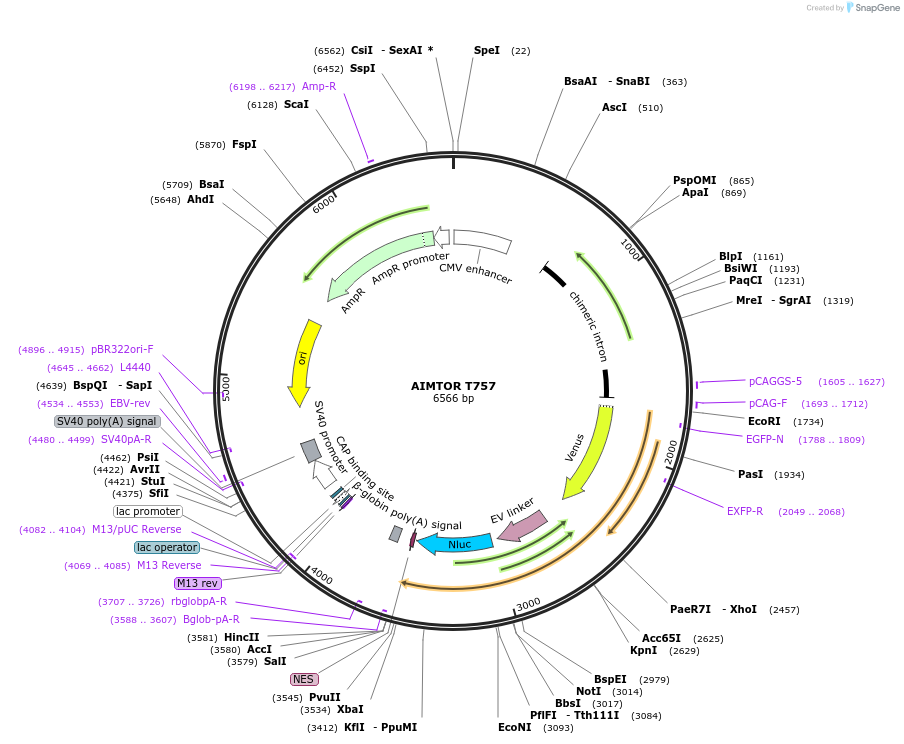
PurposeAIMTOR T757 contains a cytoplasmic mTOR Activity Reporter derived from hu ULK1 protein boxed between BRET-compatible entities (Ypet and Nanoluciferase) to measure mTOR activity in living cells
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 140828 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backboneYEN
-
Backbone manufacturerGoyet et al 2016, Scientific Reports, 6:28231. DOI: 10.1038/srep28231 and Komatsu, N et al.Molecular biology of the cell 22, 464
- Backbone size w/o insert (bp) 6529
- Total vector size (bp) 6583
-
Modifications to backboneThe plasmid coding for YEN contains a cytoplasmic Extracellular signal-regulated Kinase Activity Reporter (EKAR) boxed between BRET-compatible entities (Ypet and Nanoluciferase), pEevee-Ypet-EKARcyto-Nluc. It was designed from the optimized backbone structure of a FRET biosensor with the 116-amino-acid (SAGG)29-repeats linker (pEKAREV(3560)NES pEevee) constructed by Komatsu and colleagues (Komatsu, N. et al. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Molecular biology of the cell 22, 4647–4656, doi: 10.1091/mbc.E11-01-0072, 2011). The ECFP-coding sequence, removed by NotI/XbaI digestion, was replaced by the Nanoluciferase-coding sequence, amplified by PCR (from pNanoLuc3.2 NFKB RE) and inserted into the NotI/XbaI sites by an Infusion reaction (Clontech), The YEN plasmid served then as a backbone plasmid to construct the plasmid AIMTOR T757. To this end, we removed the sequence coding for the ERK sensor site by a BspeI/NotI restriction digest. Following a ligation with a cohesive double strand pre-annealed oligonucleotides encoding the T757 sensor peptide derived from human Ulk1, we obtained the plasmid AIMTOR T757
-
Vector typeMammalian Expression
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameAIMTOR T757
-
SpeciesH. sapiens (human)
-
Insert Size (bp)34
-
MutationThe insert comprises only a small sequence of human Ulk1 encoding for a 9 aminoacids peptide, conserved 100% in mouse Ulk1. The original serine residue phosphorylable by mTOR in this peptide (position 757 in mouse Ulk1) has been mutated to a phosphoylable threonine residue which gives higher BRET intensity variations.
-
GenBank IDNM_003565
-
Entrez GeneULK1 (a.k.a. ATG1, ATG1A, UNC51, Unc51.1, hATG1)
-
Tag
/ Fusion Protein
- NES Nuclear export signal (C terminal on insert)
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BspEI (not destroyed)
- 3′ cloning site NotI (not destroyed)
- 5′ sequencing primer CCACTACCTGAGCTACCAG
- 3′ sequencing primer GCCAGAAGTCAGATGCTCA
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
AIMTOR T757 was a gift from Anne Bonnieu (Addgene plasmid # 140828 ; http://n2t.net/addgene:140828 ; RRID:Addgene_140828) -
For your References section:
AIMTOR, a BRET biosensor for live imaging, reveals subcellular mTOR signaling and dysfunctions. Bouquier N, Moutin E, Tintignac LA, Reverbel A, Jublanc E, Sinnreich M, Chastagnier Y, Averous J, Fafournoux P, Verpelli C, Boeckers T, Carnac G, Perroy J, Ollendorff V. BMC Biol. 2020 Jul 3;18(1):81. doi: 10.1186/s12915-020-00790-8. 10.1186/s12915-020-00790-8 PubMed 32620110