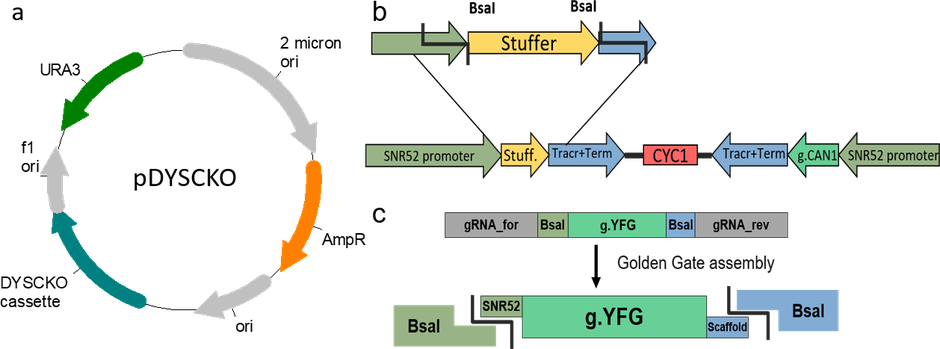
pDYSCKO
(Plasmid
#120263)
-
PurposeEmpty backbone for cloning any gRNA to be used with the canavanine co-selection system
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 120263 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonep426
- Backbone size w/o insert (bp) 5500
- Total vector size (bp) 6500
-
Modifications to backboneRemoved BsaI sites in URA3 and AmpR using silent mutations in the coding sequence.
-
Vector typeCRISPR, Synthetic Biology
-
Selectable markersURA3 ; Canavanine (after mutagenesis)
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameDYSCKO cassette
-
gRNA/shRNA sequenceATAACGGAATCCAACTGGGC
-
SpeciesS. cerevisiae (budding yeast)
- Promoter SNR52
Cloning Information
- Cloning method Gibson Cloning
- 5′ sequencing primer GCGCCGCTACAGGGCGCGT
- 3′ sequencing primer caggaaacagctatgac (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byThe gRNA expression cassette is based on p426-SNR52p-gRNA.CAN1.Y-SUP4t (#43803) from the Church lab.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
See Double Selection Enhances the Efficiency of Target-AID and Cas9-Based Genome Editing in Yeast. (PMID: 30097473) for detailed protocols for gRNA cloning and mutagenesis protocol.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pDYSCKO was a gift from Christian Landry (Addgene plasmid # 120263 ; http://n2t.net/addgene:120263 ; RRID:Addgene_120263) -
For your References section:
Double Selection Enhances the Efficiency of Target-AID and Cas9-Based Genome Editing in Yeast. Despres PC, Dube AK, Nielly-Thibault L, Yachie N, Landry CR. G3 (Bethesda). 2018 Oct 3;8(10):3163-3171. doi: 10.1534/g3.118.200461. 10.1534/g3.118.200461 PubMed 30097473