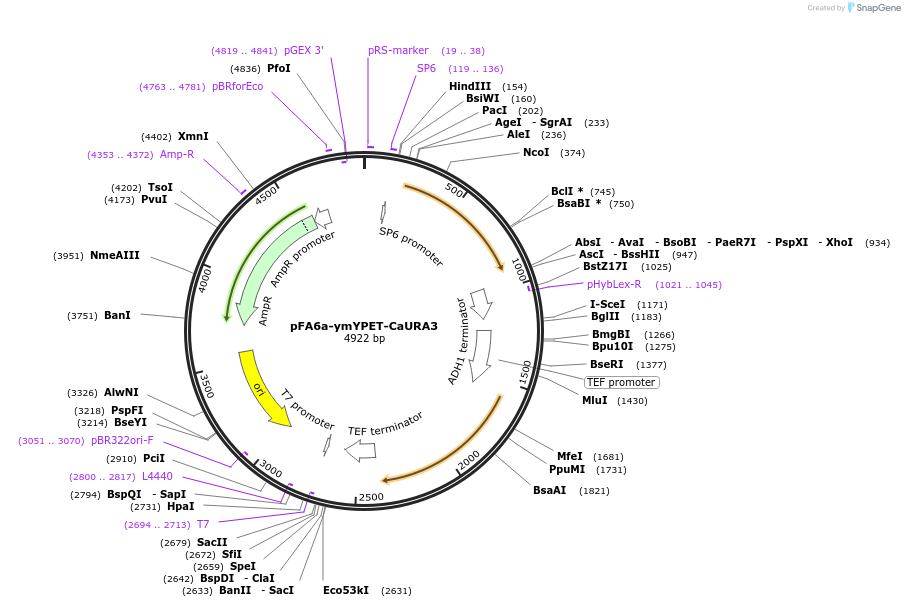
pFA6a-ymYPET-CaURA3
(Plasmid
#118460)
-
PurposeYeast tagging vector to create a gene fusion with ymYPET with CaUra3 selection. *An updated version of this plasmid is available: #168056 pFA6a-link-ymYPET-URA3.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 118460 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepFA6a
-
Vector typeYeast Expression
-
Selectable markersURA3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameymYPET
-
SpeciesSynthetic
-
MutationYPET A206K, F208S, E232L, N235D
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site unknown (unknown if destroyed)
- 3′ cloning site unknown (unknown if destroyed)
- 5′ sequencing primer Unknown
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Please note the absence of a linker region at the 5’ end of the FP. Using a FW primer upstream of the FP will introduce a stopcodon. Please use FW primers annealing on the FP itself.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pFA6a-ymYPET-CaURA3 was a gift from Bas Teusink (Addgene plasmid # 118460 ; http://n2t.net/addgene:118460 ; RRID:Addgene_118460) -
For your References section:
In vivo characterisation of fluorescent proteins in budding yeast. Botman D, de Groot DH, Schmidt P, Goedhart J, Teusink B. Sci Rep. 2019 Feb 19;9(1):2234. doi: 10.1038/s41598-019-38913-z. 10.1038/s41598-019-38913-z PubMed 30783202