-
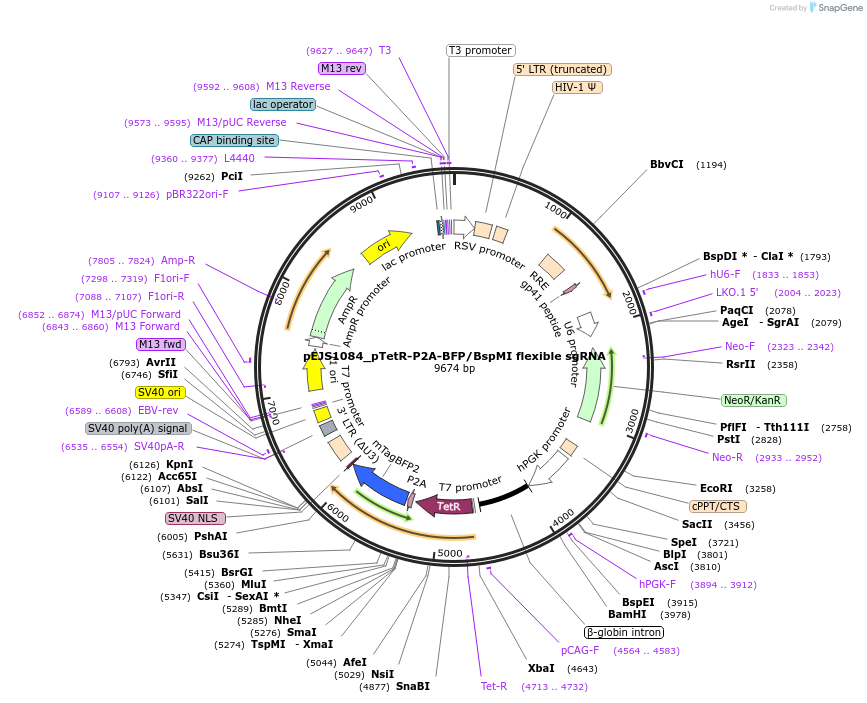
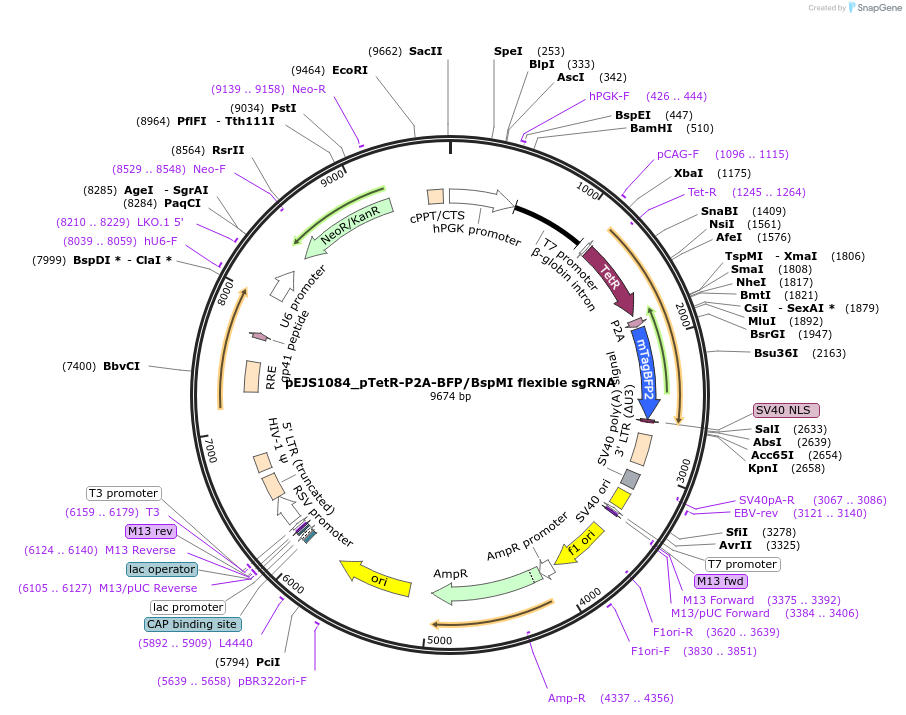
PurposeU6 driven Spy sgRNA cloning vector where guide sequences are inserted between BspMI sites. This plasmid works with Addgene #108570 for subnuclear proteomic profiling via C-BERST method.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 117687 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepLKO.1
- Backbone size w/o insert (bp) 7183
- Total vector size (bp) 9614
-
Vector typeMammalian Expression, Lentiviral, CRISPR
-
Selectable markersUse FACS to sort out BFP positive cells.
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)NEB Stable
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameKanamycin cassette
-
SpeciesBacterial origin
-
Insert Size (bp)1054
- Promoter U6
Cloning Information for Gene/Insert 1
- Cloning method Restriction Enzyme
- 5′ cloning site BspMI (destroyed during cloning)
- 3′ cloning site BspMI (destroyed during cloning)
- 5′ sequencing primer gcatatacgatacaaggctgt
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameTetR-P2A-BFP
-
SpeciesSynthetic
-
Insert Size (bp)1437
- Promoter hPGK
Cloning Information for Gene/Insert 2
- Cloning method Gibson Cloning
- 5′ sequencing primer cattctgcaagcctccgg
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pEJS1084_pTetR-P2A-BFP/BspMI flexible sgRNA was a gift from Erik Sontheimer (Addgene plasmid # 117687 ; http://n2t.net/addgene:117687 ; RRID:Addgene_117687) -
For your References section:
Adapting dCas9-APEX2 for subnuclear proteomic profiling. Gao XD, Rodriguez TC, Sontheimer EJ. Methods Enzymol. 2019;616:365-383. doi: 10.1016/bs.mie.2018.10.030. Epub 2018 Dec 10. 10.1016/bs.mie.2018.10.030 PubMed 30691651