-
Depositing Lab
-
Publication
-
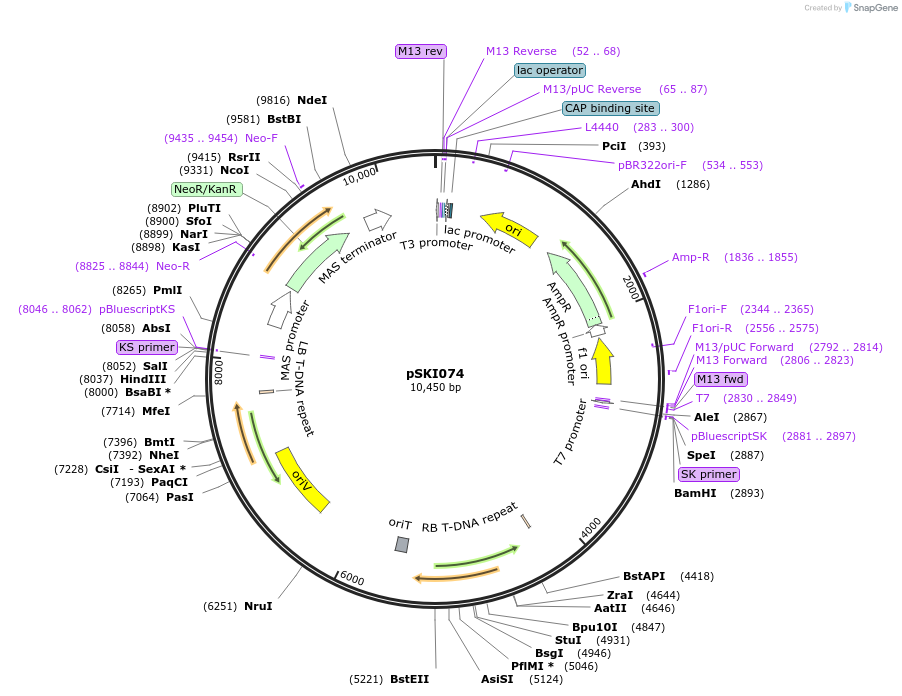
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 11571 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepPCVICEn4HPT
-
Backbone manufacturerHayashi et al, 1992
- Backbone size w/o insert (bp) 10138
-
Vector typePlant Expression ; Activation tagging
-
Selectable markerskanamycin
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Growth instructionsPlasmid grow in standard E. coli at 37 degrees, but the four enhancer repeats may be unstable in E. coli and A. tumefaciens if stored at 4'C for extended time. See comments.
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert nameCaMV 35S enhancers
-
SpeciesCauliflower mosaic virus
-
GenBank IDAF218466
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site See map and paper. (not destroyed)
- 3′ cloning site See map and paper. (not destroyed)
- 5′ sequencing primer T7
- (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byMax Planck Institute
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
The CaMV 35S enhancers in these vectors correspond to nucleotides -417 to -86 relative to the transcription start.
Recommended Agrobacterium strain is GV3101 pMP90RK. Host and helper plasmid markers are kanamycin, gentamycin and rifampicin resistance.
Plasmid selection in E. coli and in A. tumefaciens is ampicillin resistance.
Sites that leave the pBstKS+ sequences intact and cut only on one side between pBstKS+ and either left or right border can be used for plasmid rescue. Note that there are additional BssS1 sites not shown on the map.
The four enhancer repeats in the construct may be unstable in E. coli and Agrobacterium if stored at +4'C for extended time. Check by PCR with T7 (or M13-20) oligo and this one derived from the RB sequence: 5' acc cgc caa tat atc ctg 3'. This should give 1.4 kb band. Note that these oligos will not work in a transgenic plant, as the RB sequence is not transfered. Using this test, we found that after 1-2 weeks in a fridge the Agrobacterium strain loses on average one copy of a repeat, and after a month in a fridge there is only one copy left. It is a good idea to use a freshly streaked colony from a good -70'C glycerol stock for every infiltration.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pSKI074 was a gift from Detlef Weigel (Addgene plasmid # 11571 ; http://n2t.net/addgene:11571 ; RRID:Addgene_11571) -
For your References section:
Activation tagging in Arabidopsis. Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, Nguyen JT, Sato S, Wang ZY, Xia Y, Dixon RA, Harrison MJ, Lamb CJ, Yanofsky MF, Chory J. Plant Physiol. 2000 Apr . 122(4):1003-13. 10.1104/pp.122.4.1003 PubMed 10759496