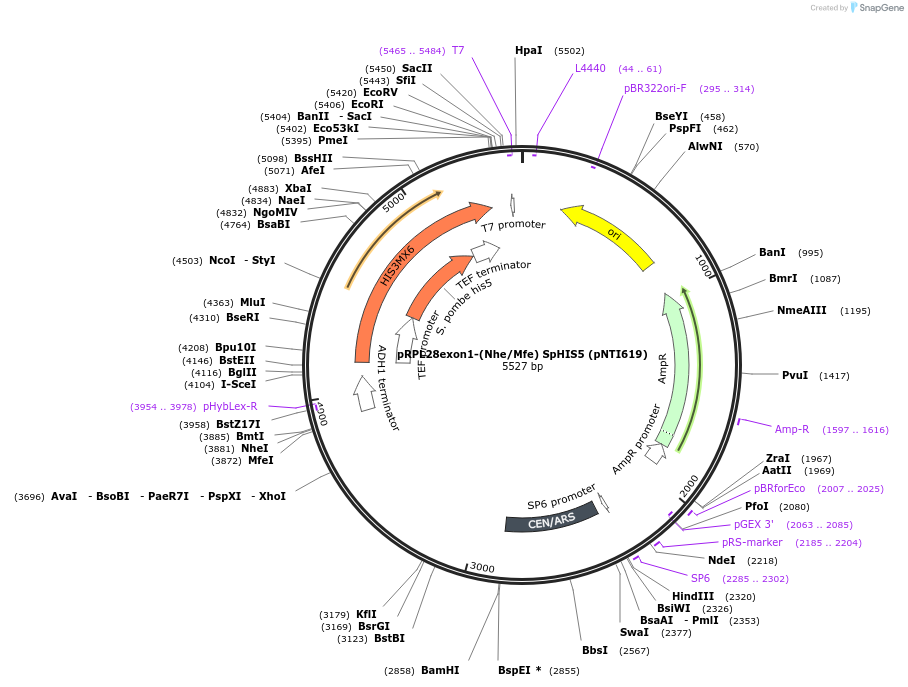
pRPL28exon1-(Nhe/Mfe) SpHIS5 (pNTI619)
(Plasmid
#115432)
-
PurposeVector for expressing RPL28 with mutagenized exon2
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 115432 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepFA6a
- Backbone size w/o insert (bp) 2500
- Total vector size (bp) 5527
-
Modifications to backboneYeast ARS/CEN
-
Vector typeYeast Expression
-
Selectable markersHIS3
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameRPL28
-
SpeciesS. cerevisiae (budding yeast)
-
Insert Size (bp)1023
-
Mutationexon 2 deleted, with MfeI / NheI cloning site
-
GenBank IDNM_001180968
-
Entrez GeneRPL28 (a.k.a. YGL103W, CYH2)
- Promoter endogenous
Cloning Information for Gene/Insert 1
- Cloning method Gibson Cloning
- 5′ sequencing primer N/A
- 3′ sequencing primer N/A (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameHIS5
-
SpeciesS. pombe (fission yeast); Candida albicans
-
Insert Size (bp)1425
- Promoter Ashbya gospii TEF1
Cloning Information for Gene/Insert 2
- Cloning method Unknown
- 5′ sequencing primer N/A
- 3′ sequencing primer N/A (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pRPL28exon1-(Nhe/Mfe) SpHIS5 (pNTI619) was a gift from Nicholas Ingolia (Addgene plasmid # 115432 ; http://n2t.net/addgene:115432 ; RRID:Addgene_115432) -
For your References section:
An Accessible Continuous-Culture Turbidostat for Pooled Analysis of Complex Libraries. McGeachy AM, Meacham ZA, Ingolia NT. ACS Synth Biol. 2019 Apr 19;8(4):844-856. doi: 10.1021/acssynbio.8b00529. Epub 2019 Apr 2. 10.1021/acssynbio.8b00529 PubMed 30908907