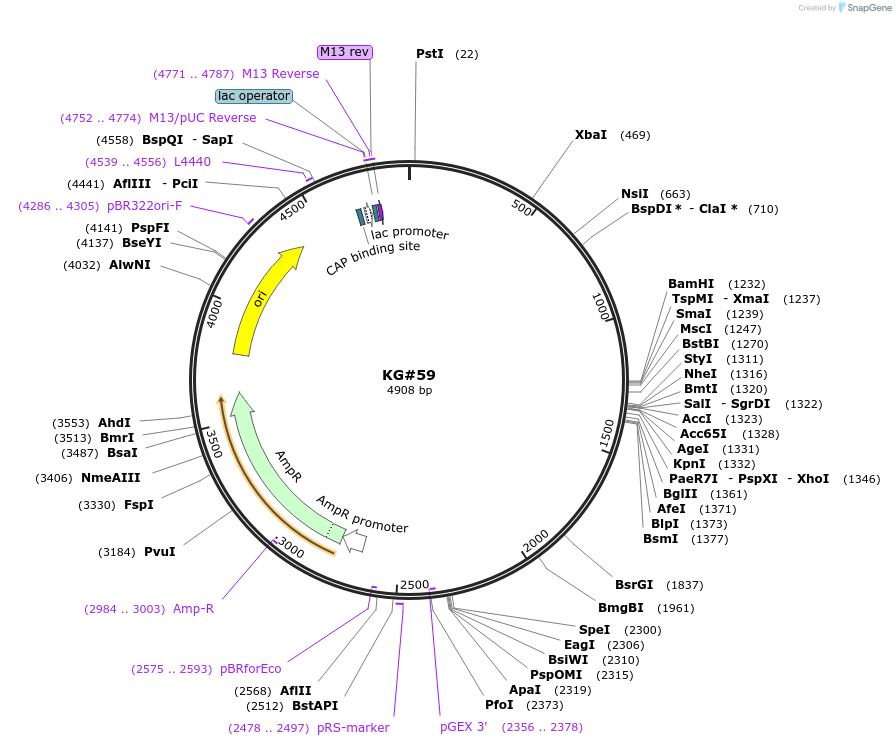
KG#59
(Plasmid
#110929)
-
PurposeExpresses proteins pan-neuronally in C. elegans (entire nervous system)
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 110929 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepUC
- Backbone size w/o insert (bp) 2632
- Total vector size (bp) 4915
-
Vector typeBacterial Expression
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert namerab-3 promoter
-
SpeciesC. elegans (nematode)
-
Insert Size (bp)1208
Cloning Information for Gene/Insert 1
- Cloning method Unknown
- 5′ sequencing primer Unknown
- (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert nameunc-54 3' control region
-
SpeciesC. elegans (nematode)
-
Insert Size (bp)940
Cloning Information for Gene/Insert 2
- Cloning method Unknown
- 5′ sequencing primer Unknown
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Unique multi-cloning sites: Nhe I, Sal I, Acc I, Kpn I/ Age I/ Xho I, Bgl II
Used Pfu Ultra polymerase and primers engineered with restriction sites to amplify the 1.2 Kb rab-3 promoter from N2 genomic DNA and cloned it into Pst I/ Bam HI cut pPD96.52 (C. elegans body wall muscle expression vector; this digestion removed the myo-3 promoter in 2 fragments and left the 3.7 kb vector). Transformed into XL1-Blue electrocompetent cells. Miniprepped 6 clones to find those with correct size insert and vector, then submitted 2 clones for sequence. Chose one clone that has correct sequence and made glycerol stock.
Features of the expression construct: This expression construct has a promoter (rab-3) that will drive strong and specific expression (of whatever gene is inserted in the MCS) in the entire nervous system of juvenile and adult animals. The boundaries of the rab-3 promoter sequence were obtained by examining pRabGFPrim3' from Mike Nonet's lab web site and comparing that to the genome sequence. Other features of the expression vector include a "decoy" 5' to the promoter to reduce non-specific expression in the gut (see Fire Lab 1995 Kit documentation for description), the unc-54 3' end including the poly A addition signal, and 3 introns (1 in 5' UTR, 1 just upstream of the unc-54 3' end, and 1 inserted in the unc-54 3' end). These introns help provide more uniform and robust expression, especially if you are expressing a cDNA with no introns as opposed to a gene with introns.
Note: this vector can be converted to a vector that fuses GFP, YFP, or CFP onto the C-terminus of the protein as follows:
Remove the ~1000 bp (Kpn I or Age I) + (Spe I or BsiW I [expensive, 55 C cutter with 50% activity at 37 C; heat inactivate 80 C] or Apa I) fragment containing the unc-54 3’ control region from this plasmid and replace it with the ~1800 bp like-digested fragment containing 3-intron S65C GFP (see pPD94.81 in Fire Lab plasmids), YFP (see pPD136.64 in Fire Lab plasmids) or CFP (see pPD136.61 in Fire Lab plasmids) + the unc-54 3’ UTR and control region. Note if Spe I is used to cut these Fire Lab vectors, you get 3 bands: 1100, 1800, and 4000 (total size 6.9 Kb). The 1800 bp band is the XFP-containing band.
For C-terminal fusions, remember to leave the stop codon off of the upstream protein. For the above-mentioned Fire Lab vectors, the reading frame for the downstream XFP relative to the Kpn I and Age I sites is as follows (triplets are the reading frame codons):
Kpn I
G GTA CCG GT
Age I
Alternatively, use Gibson Assembly/ NEBuilder to insert any genetically encoded fluorescent protein at the N- or C-terminus of your protein.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
KG#59 was a gift from Kenneth Miller (Addgene plasmid # 110929 ; http://n2t.net/addgene:110929 ; RRID:Addgene_110929) -
For your References section:
Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Schade MA, Reynolds NK, Dollins CM, Miller KG. Genetics. 2005 Feb;169(2):631-49. doi: 10.1534/genetics.104.032334. Epub 2004 Oct 16. 10.1534/genetics.104.032334 PubMed 15489510