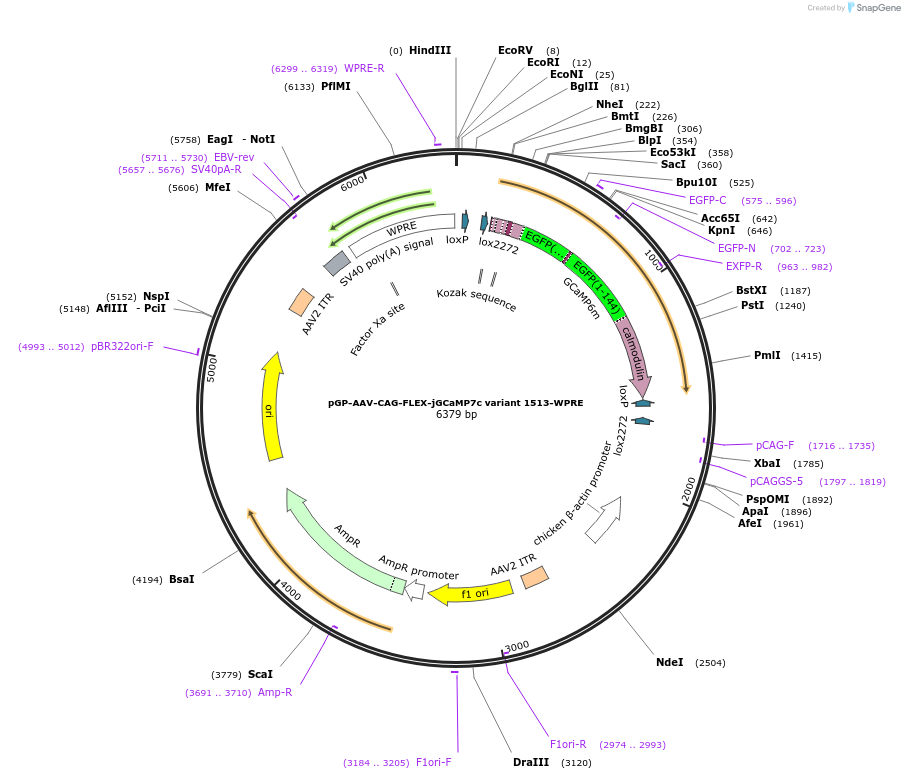
pGP-AAV-CAG-FLEX-jGCaMP7c variant 1513-WPRE
(Plasmid
#105323)
-
PurposeAAV-mediated expression of ultrasensitive protein calcium sensor under the CAG promoter, Cre-dependent expression
-
Depositing Labs
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 105323 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Want a viral vector made from this plasmid?
Make a packaging request and we'll get back to you.
Please log in to submit a packaging request.
-
SerotypeSelect serotype for details See details about
-
PricingSelect serotype and quantity $ USD for preparation of µL virus + $32 USD for plasmid.
-
How this works
- Place a request for a quantity of 2 (0.2 mL), 10 (1 mL), 25 (2.5 mL), or 50 (5 mL). Our all-inclusive pricing includes DNA production and QC.
- Addgene will quickly confirm that we can produce a high-quality prep for you.
- Track your request and place an order from within your account. Payment information must be added before we can begin processing your order.
- Receive your prep in 6–9 weeks after the MTA is approved by your organization.
- Learn more about our Packaged on Request Service.
Backbone
-
Vector backboneAAV-CAG-FLEX
-
Backbone manufacturerScott Sternson
- Backbone size w/o insert (bp) 5027
- Total vector size (bp) 6380
-
Vector typeMammalian Expression, AAV, Cre/Lox
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)NEB Stable
-
Copy numberLow Copy
Gene/Insert
-
Gene/Insert namejGCaMP7c variant 1513
-
Alt nameGCaMP3-L59Q E60P T302L R303P M378G K379S D380Y T381R R392G T412N
-
Alt nameGCaMP3 variant 1513
-
Alt nameJanelia GCaMP7
-
SpeciesR. norvegicus (rat), G. gallus (chicken); A. victoria (jellyfish)
-
Insert Size (bp)1353
- Promoter CAG
-
Tag
/ Fusion Protein
- T7 epitope, Xpress tag, 6xHis
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site BsmBI (destroyed during cloning)
- 3′ cloning site BsmBI (destroyed during cloning)
- 5′ sequencing primer GGTTCGGCTTCTGGCGTGTGACC
- (Common Sequencing Primers)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pGP-AAV-CAG-FLEX-jGCaMP7c variant 1513-WPRE was a gift from Douglas Kim & GENIE Project (Addgene plasmid # 105323 ; http://n2t.net/addgene:105323 ; RRID:Addgene_105323) -
For your References section:
High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, Macklin JJ, Chen Y, Konnerth A, Jayaraman V, Looger LL, Schreiter ER, Svoboda K, Kim DS. Nat Methods. 2019 Jul;16(7):649-657. doi: 10.1038/s41592-019-0435-6. Epub 2019 Jun 17. 10.1038/s41592-019-0435-6 PubMed 31209382