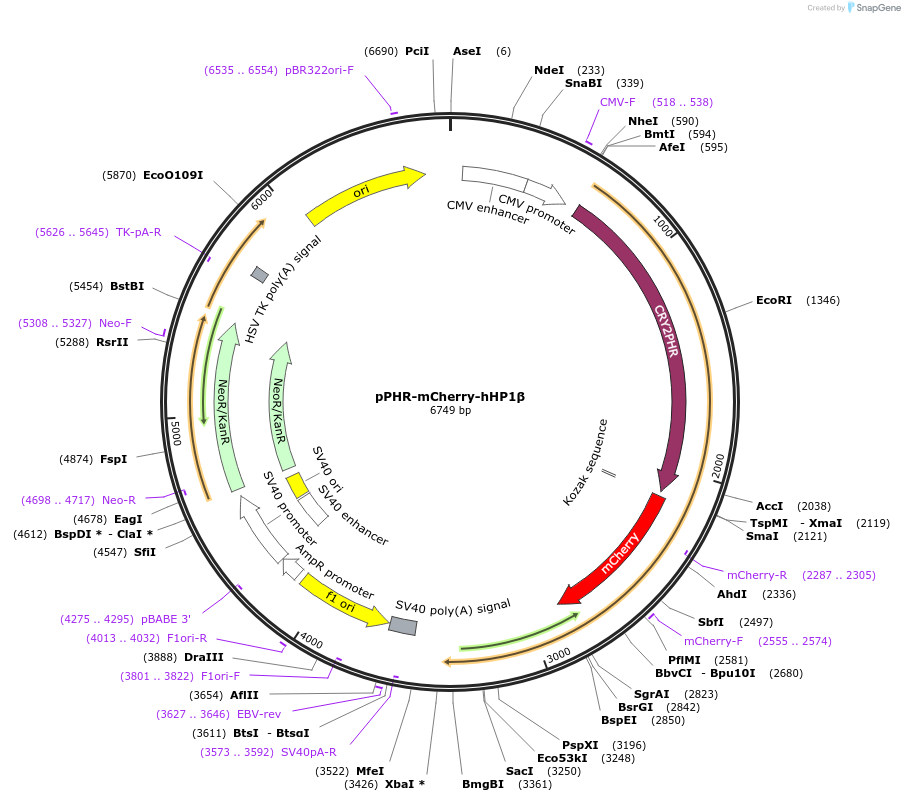
pPHR-mCherry-hHP1β
(Plasmid
#103830)
-
PurposeSoluble BLInCR effector that is recruited to 'localizer' sites upon blue light illumination
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 103830 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $89 | |
Backbone
-
Vector backbonepmCherry-N1
-
Backbone manufacturerClontech
- Backbone size w/o insert (bp) 3935
-
Vector typeMammalian Expression
-
Selectable markersNeomycin (select with G418)
Growth in Bacteria
-
Bacterial Resistance(s)Kanamycin, 50 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert
-
Gene/Insert namePHR-mCherry-hHP1β
-
Alt namePHR domain of CRY2
-
SpeciesH. sapiens (human), A. thaliana (mustard weed)
-
Insert Size (bp)2814
-
Entrez GeneCRY2 (a.k.a. AT1G04400, AT-PHH1, ATCRY2, CRYPTOCHROME 2 APOPROTEIN, F19P19.14, F19P19_14, FHA, PHH1, cryptochrome 2)
-
Entrez GeneCBX1 (a.k.a. CBX, HP1-BETA, HP1Hs-beta, HP1Hsbeta, Hp1beta, M31, MOD1, p25beta)
- Promoter CMV
Cloning Information
- Cloning method Restriction Enzyme
- 5′ cloning site XhoI (not destroyed)
- 3′ cloning site XbaI (not destroyed)
- 5′ sequencing primer CMV-F
- (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
mCherry was cut out from the backbone (XhoI/NotI cuts) and reinserted with the insert
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pPHR-mCherry-hHP1β was a gift from Karsten Rippe (Addgene plasmid # 103830 ; http://n2t.net/addgene:103830 ; RRID:Addgene_103830) -
For your References section:
Real-time observation of light-controlled transcription in living cells. Rademacher A, Erdel F, Trojanowski J, Schumacher S, Rippe K. J Cell Sci. 2017 Dec 15;130(24):4213-4224. doi: 10.1242/jcs.205534. Epub 2017 Nov 9. 10.1242/jcs.205534 PubMed 29122982