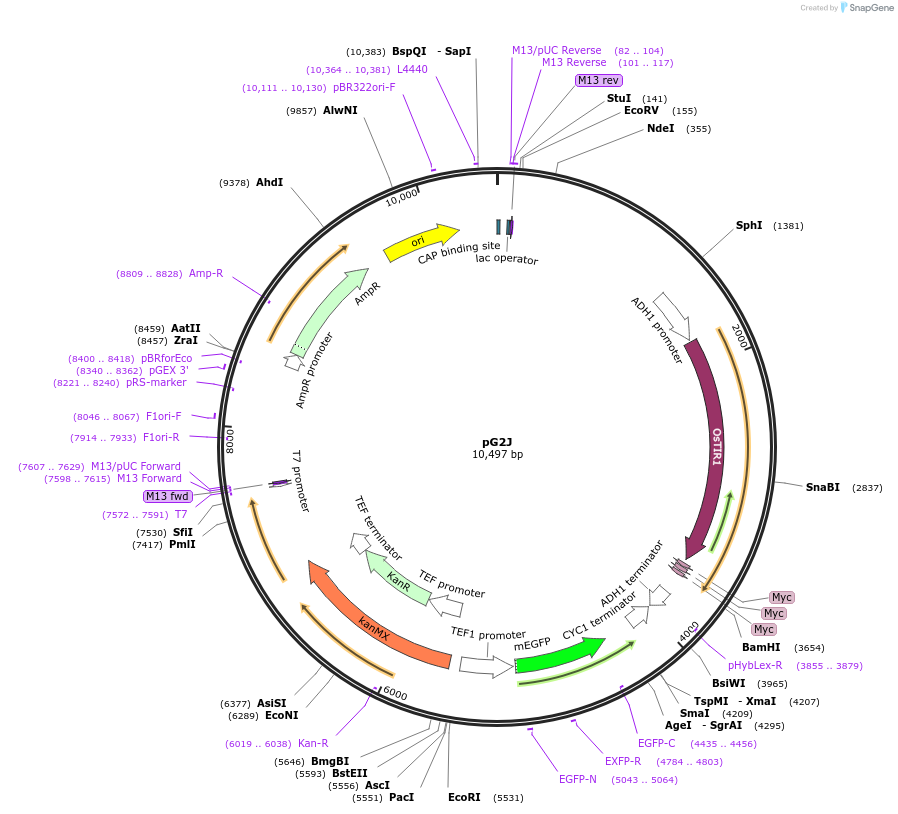
pG2J
(Plasmid
#102882)
-
PurposeExpresses OsTIR1 via the ADH1 promoter, and the mGFP-AID via the TEF1 promoter in yeast. Both cassettes are integrated in HO locus. Assembly and characterization of the auxin-inducible degradation.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 102882 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backboneHO-poly-KanMX4-HO
-
Backbone manufacturerhttps://www.ncbi.nlm.nih.gov/pmc/articles/PMC55758/
- Backbone size w/o insert (bp) 6024
- Total vector size (bp) 10502
-
Vector typeYeast Expression
-
Selectable markersKanMX4 (select with G418)
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberHigh Copy
Gene/Insert 1
-
Gene/Insert nameOsTIR1
-
Alt nameAuxin Inducible Degron
-
Alt nameAID
-
SpeciesOryza sativa
-
Insert Size (bp)2880
- Promoter ADH1
Cloning Information for Gene/Insert 1
- Cloning method Gibson Cloning
- 5′ sequencing primer GGAGTATTGTGTCATGTTCG
- 3′ sequencing primer CGAGTAAGCTTGGTACCG (Common Sequencing Primers)
Gene/Insert 2
-
Gene/Insert namemGFP-AID
-
Alt nameGFP
-
Alt nameAuxin Inducible Degron
-
Alt nametruncated AID
-
Insert Size (bp)1730
- Promoter TEF1
-
Tag
/ Fusion Protein
- AID(71-114) (C terminal on insert)
Cloning Information for Gene/Insert 2
- Cloning method Gibson Cloning
- 5′ sequencing primer ACAAGCTGGAGTACAACTAC
- 3′ sequencing primer TCTAGGGTGTCGTTAATTACC (Common Sequencing Primers)
Resource Information
-
Supplemental Documents
-
A portion of this plasmid was derived from a plasmid made byThe AID(71-114) sequence was retrieved from: Morawska, M., and Ulrich, H.D. (2013). An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351. The OsTIR1 was retrieved from: Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., and Kanemaki, M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. mGFP was retrieved from: Saraya, R., Cepińska, M.N., Kiel, J.A.K.W., Veenhuis, M., and van der Klei, I.J. (2010). A conserved function for Inp2 in peroxisome inheritance. Biochim. Biophys. Acta 1803, 617–622. The HO-poly-KanMX4-HO plasmid was retrieved from: Voth, W.P., Richards, J.D., Shaw, J.M., and Stillman, D.J. (2001). Yeast vectors for integration at the HO locus. Nucleic Acids Res. 29, E59-9.
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
This plasmid can be used to assemble and characterize the auxin inducible degron in single yeast cells.
A characterization guide can be found here:
https://www.nature.com/articles/s41598-017-04791-6
Detailed description of plasmid construction can be found in the Supplemental Experimental Procedures of the original paper:
http://www.cell.com/molecular-cell/pdfExtended/S1097-2765(16)30726-2
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pG2J was a gift from Matthias Heinemann (Addgene plasmid # 102882 ; http://n2t.net/addgene:102882 ; RRID:Addgene_102882) -
For your References section:
Autonomous Metabolic Oscillations Robustly Gate the Early and Late Cell Cycle. Papagiannakis A, Niebel B, Wit EC, Heinemann M. Mol Cell. 2017 Jan 19;65(2):285-295. doi: 10.1016/j.molcel.2016.11.018. Epub 2016 Dec 15. 10.1016/j.molcel.2016.11.018 PubMed 27989441