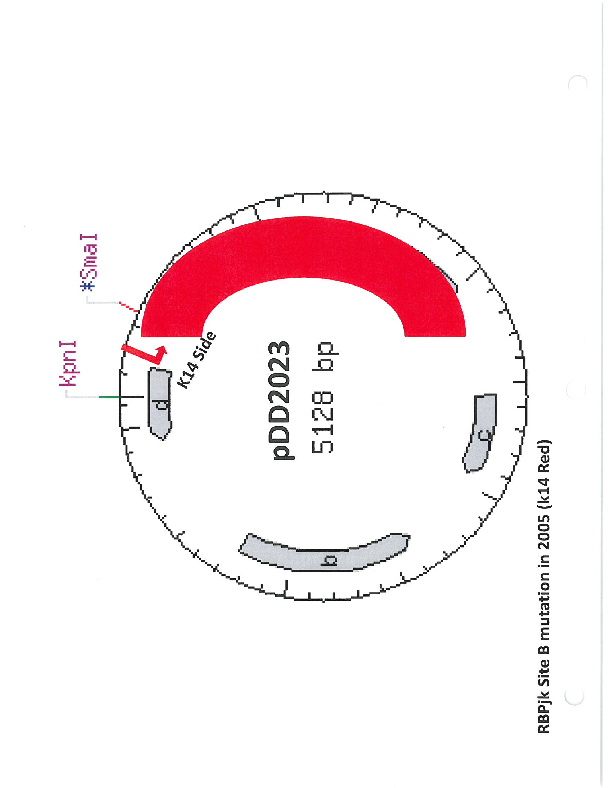
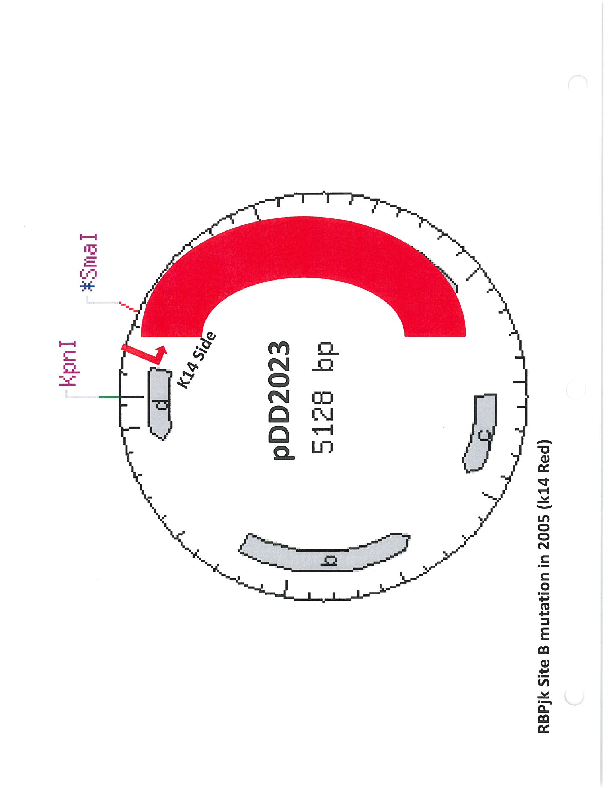
pDD2023
(Plasmid
#101367)
-
PurposeName: pCBR-RBPjkΔB. RBPjk binding site B mutation in pCBR-K14 (pDD2005).
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 101367 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonepCBR Basic
-
Backbone manufacturerPromega
- Backbone size w/o insert (bp) 5128
-
Vector typeLuciferase
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature37°C
-
Growth Strain(s)DH5alpha
-
Copy numberUnknown
Gene/Insert
-
Gene/Insert nameRBPjk binding site B mutation in pCBR-K14 (pDD2005).
-
SpeciesKSHV (viral)
-
MutationRBPjk binding site B mutation in pCBR-K14 (pDD2005).
Resource Information
-
Article Citing this Plasmid
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
Addgene's sequencing found an S168P mutation in CBRluc. This mutation is not known to affect plasmid function.
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
pDD2023 was a gift from Dirk Dittmer (Addgene plasmid # 101367 ; http://n2t.net/addgene:101367 ; RRID:Addgene_101367) -
For your References section:
Quantitative analysis of the bidirectional viral G-protein-coupled receptor and lytic latency-associated nuclear antigen promoter of Kaposi's sarcoma-associated herpesvirus. Hilton IB, Dittmer DP. J Virol. 2012 Sep;86(18):9683-95. doi: 10.1128/JVI.00881-12. Epub 2012 Jun 27. 10.1128/JVI.00881-12 PubMed 22740392