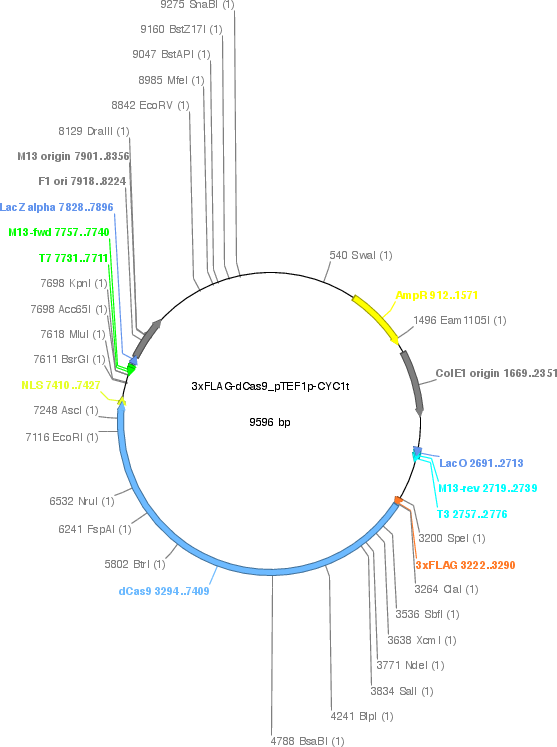
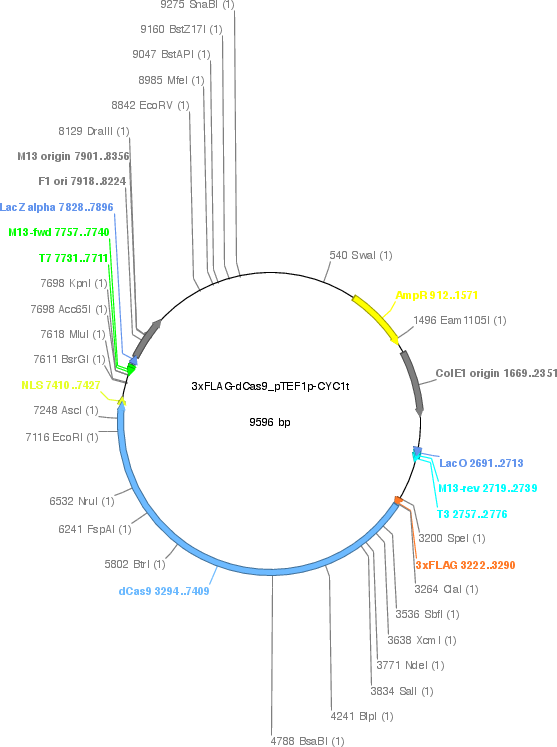
3xFLAG-dCas9/pTEF1p-CYC1t
(Plasmid
#62190)
-
PurposeExpresses 3xFLAG-dCas9 in budding yeast for enChIP analysis to purify specific genomic regions of interest.
-
Depositing Lab
-
Sequence Information
Ordering
| Item | Catalog # | Description | Quantity | Price (USD) | |
|---|---|---|---|---|---|
| Plasmid | 62190 | Standard format: Plasmid sent in bacteria as agar stab | 1 | $85 | |
Backbone
-
Vector backbonep414
- Backbone size w/o insert (bp) 5385
- Total vector size (bp) 9596
-
Vector typeYeast Expression, CRISPR
-
Selectable markersTRP1
Growth in Bacteria
-
Bacterial Resistance(s)Ampicillin, 100 μg/mL
-
Growth Temperature30°C
-
Growth Strain(s)NEB Stable
-
Copy numberLow Copy
Gene/Insert
-
Gene/Insert name3xFLAG-dCas9
-
SpeciesSynthetic; S. pyogenes
-
Insert Size (bp)4212
-
Mutationhuman codon-optimized, D10A, H840A
- Promoter TEF1 promoter
-
Tags
/ Fusion Proteins
- 3xFLAG (N terminal on insert)
- NLS (C terminal on insert)
Cloning Information
- Cloning method Unknown
- 5′ sequencing primer T3 promoter
- 3′ sequencing primer T7 promoter (Common Sequencing Primers)
Resource Information
-
A portion of this plasmid was derived from a plasmid made byGeorge Church (Addgene plasmid 43802)
Terms and Licenses
-
Academic/Nonprofit Terms
-
Industry Terms
- Not Available to Industry
Trademarks:
- Zeocin® is an InvivoGen trademark.
Depositor Comments
This plasmid is compatible with gRNA expression plasmids for budding yeast, such as Addgene Plasmid #43803 ( http://www.addgene.org/43803 ).
For more information on Fujii Lab CRISPR Plasmids please refer to: http://www.addgene.org/crispr/fujii/
These plasmids were created by your colleagues. Please acknowledge the Principal Investigator, cite the article in which the plasmids were described, and include Addgene in the Materials and Methods of your future publications.
-
For your Materials & Methods section:
3xFLAG-dCas9/pTEF1p-CYC1t was a gift from Hodaka Fujii (Addgene plasmid # 62190 ; http://n2t.net/addgene:62190 ; RRID:Addgene_62190) -
For your References section:
An enChIP system for the analysis of genome functions in budding yeast. Fujii H, Fujita T. Biol Methods Protoc. 2022 Oct 17;7(1):bpac025. doi: 10.1093/biomethods/bpac025. eCollection 2022. 10.1093/biomethods/bpac025 PubMed 36325175